tpj12229-sup-0039-Legend
advertisement

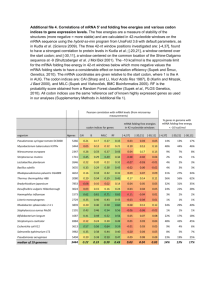
SUPPLEMENTARY LEGENDS Figure S1. GO term enrichment in the signature genes of the foliar and husk leaf samples. GO terms in FP/HP (purple segments), FP3/4 & HP/3/4 (orange segments), FP5/HP5 (yellow segments), FI/HI (light green segments) and FE/HE (dark green segments) samples. GO terms that are significantly enriched in the signature genes of both foliar (F) and husk (H) samples are presented in black text; foliar only – blue; husk only – red. Figure S2. Pathway enrichment in the signature genes of foliar and husk leaf samples. (a, b) Terms enriched in both foliar (F) and husk (H) samples are presented in black text; foliar only – blue; husk only – red. FI/HI are depicted as light green segments (a); FE/HE as dark green segments (b). Figure S3. Maximum likelihood phylogenetic tree of auxin response factor proteins. Values at nodes indicate approximate SH-like support values. Cartoons next to sequences depict the domain organisation of the predicted protein sequence. Sequence is indicated by the black line and domains are coloured according to legend in bottom left corner. Where the Arabidopsis gene has been named the name is given next to the sequence. “****” denotes genes that are present in Figure 4a. Figure S4. Maximum likelihood phylogenetic tree of AUX/IAA family proteins. Values at nodes indicate approximate SH-like support values. Cartoons next to sequences depict the domain organisation of the predicted protein sequence. Sequence is indicated by the black line and domains are coloured according to legend in bottom left corner. Where the Arabidopsis gene has been named the name is given next to the sequence. “****” denotes genes that are present in Figure 4a. Figure S5. Maximum likelihood phylogenetic tree of zinc finger family proteins. Values at nodes indicate approximate SH-like support values. Cartoons next to sequences depict the domain organisation of the predicted protein sequence. Sequence is indicated by the black line and domains 1 are coloured according to legend in bottom left corner. Where the Arabidopsis gene has been named, the name is given next to the sequence. Alternatively the sequence is labelled “not named.” “****” denotes genes that are present in Figure 4a. Figure S6. Maximum likelihood phylogenetic tree of C2H2 family transcription factors. Values at nodes indicate approximate SH-like support values. Cartoons next to sequences depict the domain organisation of the predicted protein sequence. Sequence is indicated by the black line and domains are coloured according to legend in bottom left corner. Where the Arabidopsis gene has been named the name is given next to the sequence. Alternatively, the sequence is labelled “not named.” “****” denotes genes that are present in Figure 4a. Figure S7. Maximum likelihood phylogenetic tree of MYB family transcription factors. Values at nodes indicate approximate SH-like support values. Cartoons next to sequences depict the domain organisation of the predicted protein sequence. Sequence is indicated by the black line and domains are coloured according to legend in bottom left corner. Where the Arabidopsis gene has been named, the name is given next to the sequence. “****” denotes genes that are present in Figure 4a. Figure S8. Pairwise sample correlations and quantitative PCR validation of RNAseq abundance estimates. (a, b) Spearman ranked correlation coefficient for the global mRNA abundance for all pairwise sample comparisons. Correlation shown as a heatmap (strongest correlation in red, weakest correlation black) (a), numerical values for (a) are provided in (b). Both sequencing replicates for each of the 10 samples are shown. (c) Correlation between qPCR and mRNA abundance estimate from RNAseq (r2 = 0.867, p < 5.6 10-17). Table S1. Putative negative regulators of Kranz anatomy in maize. Rice ortholog ID and transcript abundance in rice seedlings as determined by read values in GEO dataset accession number GSE33265. 2 Table S2. Primer pairs used for qRT-PCR. Data S1. mRNA abundance estimates and annotations for FP signature genes. Data S2. mRNA abundance estimates and annotations for FP3/4 signature genes. Data S3. mRNA abundance estimates, annotations and enrichment test results for GO terms, MaizeCyc pathways, Pfam domains and MapMan terms for FP5 signature genes. Data S4. mRNA abundance estimates, annotations and enrichment test results for GO terms, MaizeCyc pathways, Pfam domains and MapMan terms for FI signature genes. Data S5. mRNA abundance estimates, annotations and enrichment test results for GO terms, MaizeCyc pathways, Pfam domains and MapMan terms for FE signature genes. Data S6. mRNA abundance estimates and annotations for HP signature genes. Data S7. mRNA abundance estimates and annotations for HP3/4 signature genes. Data S8. mRNA abundance estimates, annotations and enrichment test results for GO terms, MaizeCyc pathways, Pfam domains and MapMan terms for HP5 signature genes. Data S9. mRNA abundance estimates, annotations and enrichment test results for GO terms, MaizeCyc pathways, Pfam domains and MapMan terms for HI signature genes. Data S10. mRNA abundance estimates, annotations and enrichment test results for GO terms, MaizeCyc pathways, Pfam domains and MapMan terms for HE signature genes. Data S11. mRNA abundance estimates, annotations and enrichment test results for GO terms, Pfam domains and MapMan terms for FD1 profile genes. Data S12. mRNA abundance estimates, annotations and enrichment test results for GO terms and MapMan terms for FD2 profile genes. Data S13. mRNA abundance estimates, annotations and enrichment test results for GO terms, MaizeCyc pathways, Pfam domains and MapMan terms for FD3 profile genes. Data S14. mRNA abundance estimates, annotations and enrichment test results for GO terms, Pfam domains and MapMan terms for FA1 profile genes. Data S15. mRNA abundance estimates, annotations and enrichment test results for GO terms, MaizeCyc pathways, Pfam domains and MapMan terms for FA2 profile genes. 3 Data S16. mRNA abundance estimates, annotations and enrichment test results for GO terms, MaizeCyc pathways, Pfam domains and MapMan terms for FA3 profile genes. Data S17. mRNA abundance estimates, annotations and enrichment test results for GO terms, MaizeCyc pathways, Pfam domains and MapMan terms for FN profile genes. Data S18. mRNA abundance estimates, annotations and enrichment test results for GO terms, Pfam domains and MapMan terms for HD1 profile genes. Data S19. mRNA abundance estimates, annotations and enrichment test results for GO terms, Pfam domains and MapMan terms for HD2 profile genes. Data S20. mRNA abundance estimates, and annotations for HD3 profile genes. Data S21. mRNA abundance estimates, annotations and enrichment test results for GO terms, MaizeCyc pathways, Pfam domains and MapMan terms for HA1 profile genes. Data S22. mRNA abundance estimates, annotations and enrichment test results for GO terms, Pfam domains and MapMan terms for HA2 profile genes. Data S23. mRNA abundance estimates, annotations and enrichment test results for GO terms, MaizeCyc pathways, Pfam domains and MapMan terms for HA3 profile genes. Data S24. mRNA abundance estimates, annotations and enrichment test results for GO terms, Pfam domains and MapMan terms for for HN profile genes. Data S25. Manual annotation of transcription factors in all descending and ascending profiles. Data S26. mRNA abundance estimates, annotations and enrichment test results for GO terms, Pfam domains and MapMan terms for 283 putative positive regulators of Kranz. Data S27. mRNA abundance estimates, annotations and enrichment test results for GO terms, Pfam domains and MapMan terms for 142 putative negative regulators of Kranz. Data S28. Summary data for all detected genes showing maize ID, EntrezGene classification, rpkm values for replicates of all 10 samples, presence in signature gene set (if relevant) plus affiliation with specific foliar and husk profiles. 4