References - WordPress.com
advertisement

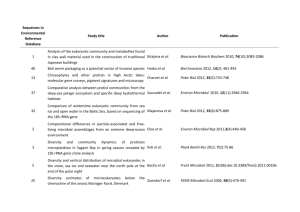
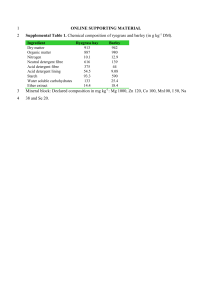
References Abed RM, Safi NM, Köster J et al (2002) Microbial diversity of a heavily polluted microbial mat and its community changes following degradation of petroleum compounds. Appl Environ Microbiol 68:1674–1683. Al Hasan RH, Sorkhoh NA, Al Bader D et al (1994) Utilization of hydrocarbons by cyanobacteria from microbial mats on oily coasts of the Gulf. Appl Microbiol Biotechnol 41:615–619. Al-Hasan RH, Al-Bader DA, Sorkhoh NA et al (1998) Evidence for n-alkane consumption and oxidation by filamentous cyanobacteria from oil contaminated coasts of the Arabian Gulf. Marine Biol 130:521–527. Altschul, S.F., Madde, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. Antal TK, Lindblad P (2005) Production of H2 by sulphur-deprived cells of the unicellular cyanobacteria Gloeocapsa alpicola and Synechocystis sp. PCC 6803 during dark incubation with methane or at various extracellular pH. J Appl Microbiol 98:114–120. Badger, M.R., Price, G.D. (2003) CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot 54: 609-622. Battistuzzi FU, Feijao A, Hedges SB (2004) A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol Bio. 4:44. Bentley, S.D., Parkhill, J. (2004) Comparative genome structure of prokaryotes. Annu Rev Genet 38: 771–91. Boucher, Y., Douady, C.J., Papke, R.T., Walsh, D.A., Boudreau, M.E.R., Nesbo., C.L., Case, R.J., Doolittle, W.F. (2003) Lateral gene transfer and the origin of prokaryotic groups. Annu Rev Genet 37: 283–328. Burja, A.M., Dhamwichukorn, S., Wright, P.C. (2003) Cyanobacterial post genomic research and systems biology. Trends Biotechnol 21: 504-511. Caldwell SL, Laidler JR, Brewer EA et al (2008) Anaerobic oxidation of methane: mechanisms, bioenergetics, and the ecology of associated microorganisms. Environ Sci Technol 42:6791– 6799. 30 Caldwell, S.L., Laidler, J.R., Brewer, E.A., Eberly, J.O., Sandborgh, S.C., Colwell, F.S. (2008) Anaerobic oxidation of methane: mechanisms, bioenergetics, and the ecology of associated microorganisms. Environ Sci Technol 42: 6791–6799. Cavalier-Smith, T. (2006) Cell evolution and Earth history: stasis and revolution. Philos Trans R Soc Lond B Biol Sci 361: 969-1006. Chaillan F, Gugger M, Saliot A et al (2005). Role of cyanobacteria in the biodegradation of crude oil by a tropical cyanobacterial mat. Chemosphere 62:1574–1582. Chellapandi P (2011a) Computational studies on enzyme-substrate complexes of methanogenesis for revealing their substrate binding affinities to direct the reverse reactions. Prot Pept Lett (In Press). Chellapandi P (2011b) In silico description of cobalt and tungsten assimilation systems in the genomes of methanogens. Syst Synt Biol. DOI 10.1007/s11693-011-9087-2 Chellapandi P (2011c) Molecular evolution of methanogens based on their metabolic facets. Front Biol 6:490-503. Chellapandi P, Dhivya C (2010) Overview of microbial metabolomics: A special insight to cyanobacterial methylotrophy. J Adv Dev Res 1:59–73. Chistoserdova L (2011) Modularity of methylotrophy, revisited. Environ Microbiol 13:26032622. Chistoserdova L, Kalyuzhnaya MG, Lidstrom ME (2009) The expanding world of methylotrophic metabolism. Annu Rev Microbiol 63:477–499. Chistoserdova, L., Jenkins, C., Kalyuzhnaya, M., Marx, C.J., Lapidus, A., Vorholt, J.A., Staley, J.T., Lidstrom, M.E. (2004) The enigmatic Planctomycetes may hold a key to the origins of methanogenesis and methylotrophy. Mol Biol Evol 21: 1234-1241. Chistoserdova, L., Vorholt, J.A., Lidstrom, M.E. (2005) A genomic view of methane oxidation by aerobic bacteria and anaerobic archaea. Genome Biol 6: 208. Dam, P., Zhengehang, S., Olman, V., Xu, Y. (2004) In silico construction of the carbon fixation pathway in Synechococcus sp. WH8102. IEEE Comput Syst Bioinformatics Conf. de Marsac, N.T., Houmard, J. (2006) Adaptation of cyanobacteria to environmental stimuli: new steps towards molecular mechanisms. FEMS Microbiol Lett 104: 119-189. Dubey SK, Dubey J, Mehra S et al (2011) Potential use of cyanobacterial species in bioremediation of industrial effluents. African J Biotechnol 10:1125–1132. 31 Figge, R.M., Cassier-Chauvat, C., Chauvat, F., Cerff, R. (2001) Characterization and analysis of an NAD(P)H dehydrogenase transcriptional regulator critical for the survival of cyanobacteria facing inorganic carbon starvation and osmotic stress. Mol Microbiol 39: 455-468. Forster, P., Ramaswamy, V., Artaxo, P., Berntsen, T., Betts, R., Fahey, D.W., Haywood, J., Lean, J., Lowe, D.C., Myhre, G., Nganga, J., Prinn, R., Raga, G., Schulz, M., Van Dorland, R.. (2007) Changes in atmospheric constituents and in radiative forcing. In: Climate change 2007. Solomon, S., Qin, D., Manning, M., Chen, Z., Marquis, M., Averyt, K.B., Tignor, M., Miller, H.L. (eds.). The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, United Kingdom and New York, USA. Garcia-Vallve, S., Guzman, E., Montero, M.A., Romeu, A. (2003) HGT-DB: a database of putative horizontally transferred genes in prokaryotic complete genomes. Nucleic Acids Res 31: 187-189. Gerdes, S.Y., Kurnasov, O.V., Shatalin, K., Polanuyer, B., Sloutsky, R., Vonstein, V., Overbeek, R., Osterman, A.L. (2006) Comparative genomics of NAD biosynthesis in cyanobacteria. J Bacteriol 188: 3012-3023. Gibson, J.L., Tabita, F.R. (1996) The molecular regulation of the reductive pentose phosphate pathway in proteobacteria and cyanobacteria. Arch Microbiol 166: 141-150. Ginsburg, H. (2009) Caveat emptor: limitations of the automated reconstruction of metabolic pathways in Plasmodium. Trends Parasitol. 2009 Jan;25(1):37-43. Gupta, R.S. (2000) The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol Rev 24: 367-402. Hakemian, A.S., Rosenzweig, A.C. (2007) The Biochemistry of Methane Oxidation. Annu Rev Biochem 76: 223-241. Higgins, C.F. (2001) ABC transporters: physiology, structure and mechanism - an overview. Res Microbiol 152: 205-210. Janssen DB, Pries F, van der Ploeg JR (1994) Genetics and biochemistry of dehalogenating enzymes. Annu Rev Microbiol 48:163–191. Kimura, M. (1983) The neutral theory of molecular evolution. Cambridge: Cambridge Univ. Press 32 Kirkwood AE, Nalewajko C, Fulthorpe RR (2006) The effects of cyanobacterial exudates on bacterial growth and biodegradation of organic contaminants. Microb Ecol 51:4–12. Knittel K, Boetius A (2009) Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol 63:311–334. Koonin, E.V. (2005) Orthologs, Paralogs, and Evolutionary Genomics. Annu Rev Genet 39: 309–38. Kuritz T, Wolk CP (1995) Use of filamentous cyanobacteria for biodegradation of organic pollutants. Appl Environ Microbiol 61:234–238. Lakshmi, P.T.V. (2007) An insight into cyanobacterial genomics - a perspective. Bioinformation 2: 8–11. Laskowski, R.A., Watson, J.D., Thornton, J.M. (2005). ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res 33: W89-W93. Leahy JG, Colwell RR (1990) Microbial degradation of hydrocarbons in the environment. Microbiol Rev 54:305–315. Marchler-Bauer, A., Anderson, J.B., Chitsaz, F., Derbyshire, M.K., DeWeese-Scott, C., Fong, J.H., Geer, L.Y., Geer, R.C., Gonzales, N.R., Gwadz, M., He, S., Hurwitz, D.I., Jackson, J.D., Ke, Z., Lanczycki, C.J., Liebert, C.A., Liu, C., Lu, F., Lu, S., Marchler, G.H., Mullokandov, M., Song, J.S., Tasneem, A., Thanki, N., Yamashita, R.A., Zhang, D., Zhang, N., Bryant, S.H. (2009) CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res 37: 205-210. Marchler-Bauer, A., Bryant, S.H. (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32: 327-331. Markov, A.V., Zakharov, I.A. (2009) Evolution of gene orders in genomes of cyanobacteria. Russian J Genet 45: 906–916. Markowitz, V.M., Ivanova, N., Szeto, E., Palaniappan, K., Chu, K., Dalevi, D., Chen, I.M., Grechkin, Y., Dubchak, I., Anderson, I., Lykidis, A., Mavromatis, K., Hugenholtz, P., Kyrpides, N.C. (2008) IMG/M: a data management and analysis system for metagenomes, Nucleic Acids Res 36: D534 - D538. Mattes TE, Alexander AK, Coleman NV (2010) Aerobic biodegradation of the chloroethenes: pathways, enzymes, ecology and evolution. FEMS Microbiol Rev 34:445–475. 33 Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A.C., Kanehisa, M. (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35: W182-W185. Nair, S., Finkel, S.E. (2004) Dps protects cells against multiple stresses during stationary phase. J Bacteriol 186: 4192-4198. Penny C, Vuilleumier S, Bringel F (2010) Microbial degradation of tetrachloromethane: mechanisms and perspectives for bioremediation. FEMS Microbiol Ecol 74:257–275. Prasanna, R., Kumar, V., Kumar, S., Yadav, A.K., Tripathi, U., Singh, A.K., et al. (2002) Methane production in rice soil is inhibited by cyanobacteria. Microbiol Res 157: 1–6. Radwan SS, Al-Hasan RH (2001) Potential application of coastal biofilm-coated gravel particles for treating oily waste. Aquatic Microbial Ecol 23:113–117. Raghukumar, C., Vipparty, V., David, J.J., Chandramohan, D. (2001) Degradation of crude oil by marine cyanobacteria. Appl Microbiol Biotechnol 57: 433-436. Raymond, J. (2005) The Evolution of biological carbon and nitrogen cycling - a genomic perspective. Rev Mineral Geochem 59: 211-231. Ritchie, R.J., Islam, N. (2001) Permeability of methylamine across the membrane of a cyanobacterial cell. New Phytologist 152: 203-211. Rocap, G., Larimer, F.W., Lamerdin, J., Malfatti, S., Chain, P., Ahlgren, N.A., Arellano, A., Coleman, M., Hauser, L., Hess, W.R., Johnson, Z.I., Land, M., Lindell, D., Post, A.F., Regala, W., Shah, M., Shaw, S.L., Steglich, C., Sullivan, M.B., Ting, C.S., Tolonen, A., Webb, E.A., Zinser, E.R., Chisholm, S.W. (2003) Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424: 1042–1047. Rocha, E.P.C. (2008) The organization of the bacterial genome. Annu Rev Genet 42: 211-233. Rojo F (2009) Degradation of alkanes by bacteria. Environ Microbiol 11:2477–2490. Rudi, K., Skulberg, O.M., Jakobsen, K. (1998) Evolution of cyanobacteria by exchange of genetic material among phyletically related strains. J Bacteriol 180: 3453–3461. Schafer H, Myronova N, Boden R (2010) Microbial degradation of dimethylsulphide and related C1-sulphur compounds: organisms and pathways controlling fluxes of sulphur in the biosphere. J Exp Bot 61:315–334. Schwartzman, D., Caldeira, K., Pavlov, A. (2008) Cyanobacterial emergence at 2.8 Gya and greenhouse feedbacks. Astrobiol 8: 187-203. 34 Sheridan, P.P., Freeman, K.H., Brenchley, J.E. (2003) Estimated minimal divergence times of the major bacterial and archaeal phyla. Geomicrobiol J 20: 1–14. Sugaya, N., Murakami, H., Satoh, M. (2003) Gene distribution patterns on cyanobacterial genomes. Genome Informatics 14: 561-562. Tamagnini, P., Axelsson, R., Lindberg, P., Oxelfelt, F., Wunschiers, R., Lindblad, P. (2002) Hydrogenases and hydrogen metabolism of cyanobacteria. Microbiol Mol Biol Rev 66: 1-20. Tamura, K., Dudley, J., Nei, M., Kumar, S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596-1599. Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., Higgins, D.G. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. Tian, W., Skolnick, J., (2003) How Well is Enzyme Function Conserved as a Function of Pairwise Sequence Identity?. J Mol Biol 333: 863–882. Tomitani, A., Knoll, A.H., Cavanaugh, C.M., Ohno, T. (2006) The evolutionary diversification of cyanobacteria: Molecular–phylogenetic and paleontological perspectives. Proc Natl Acad Sci USA 103: 5442–5447. Uchiyama, I. (2006) Hierarchical clustering algorithm for comprehensive orthologous-domain classification in multiple genomes. Nucleic Acids Res 34: 647–658. Vorholt, J.A. (2002) Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch Microbiol 178: 239–49. Wolfe, R.S., Higgins, L.J. (1979) Microbial biochemistry of methane - a study in contrasts. Int Rev Biochem 21: 267-353. Woodger, F.J., Badger, M.R., Price, G.D. (2003) Inorganic carbon limitation induces transcripts encoding components of the CO(2)-concentrating mechanism in Synechococcus sp. PCC7942 through a redox-independent pathway. Plant Physiol 133: 2069-2080. Wu ML, Ettwig KF, Jetten MS et al (2011) A new intra-aerobic metabolism in the nitritedependent anaerobic methane-oxidizing bacterium Candidatus 'Methylomirabilis oxyfera'. Biochem Soc Trans 39:243–248. Zhu G, Jetten MS, Kuschk P et al (2010) Potential roles of anaerobic ammonium and methane oxidation in the nitrogen cycle of wetland ecosystems. Appl Microbiol Biotechnol 86:1043– 1055. 35 Zhuang, Q., Melack, J.M., Zimov, S., Walter, K.M., Butenhoff, C.L., Khalil, M.A.K. (2009) Transactions, American Geophysical Union 90: 37-44. 36