Supplement 1. Model variables used to create cost surfaces for
advertisement
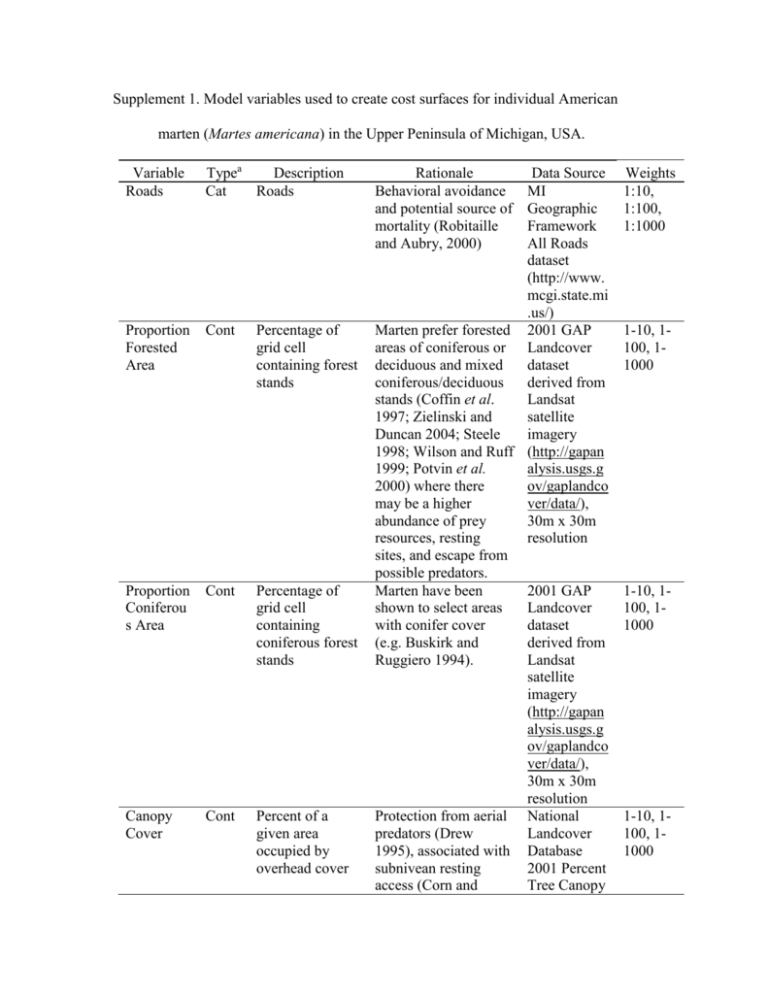
Supplement 1. Model variables used to create cost surfaces for individual American marten (Martes americana) in the Upper Peninsula of Michigan, USA. Variable Roads Typea Description Cat Roads Proportion Forested Area Cont Percentage of grid cell containing forest stands Proportion Coniferou s Area Cont Percentage of grid cell containing coniferous forest stands Canopy Cover Cont Percent of a given area occupied by overhead cover Rationale Behavioral avoidance and potential source of mortality (Robitaille and Aubry, 2000) Data Source MI Geographic Framework All Roads dataset (http://www. mcgi.state.mi .us/) Marten prefer forested 2001 GAP areas of coniferous or Landcover deciduous and mixed dataset coniferous/deciduous derived from stands (Coffin et al. Landsat 1997; Zielinski and satellite Duncan 2004; Steele imagery 1998; Wilson and Ruff (http://gapan 1999; Potvin et al. alysis.usgs.g 2000) where there ov/gaplandco may be a higher ver/data/), abundance of prey 30m x 30m resources, resting resolution sites, and escape from possible predators. Marten have been 2001 GAP shown to select areas Landcover with conifer cover dataset (e.g. Buskirk and derived from Ruggiero 1994). Landsat satellite imagery (http://gapan alysis.usgs.g ov/gaplandco ver/data/), 30m x 30m resolution Protection from aerial National predators (Drew Landcover 1995), associated with Database subnivean resting 2001 Percent access (Corn and Tree Canopy Weights 1:10, 1:100, 1:1000 1-10, 1100, 11000 1-10, 1100, 11000 1-10, 1100, 11000 Raphael 1992), and prey (Thompson and Colgan 1994) Fisher Harvest Density a Cont Density of harvested fishers dataset (http://www. mrlc.gov/nlc d2001.php), 30m x 30m resolution Fisher may predate on Derived from 1-10, 1marten (Raine 1987) Michigan 100, 1and represent a source Department 1000 of indirect competition of Natural for food resources, Resources particularly small fisher harvest mammal prey (Krohn locations et al. 1997) and during 2000denning sites (Clem 2004 and 1977). ESRI ArcGIS 9.3 Kernel Density Tool Cont = Continuous, Cat = Categorical. 5 Buskirk SW, Ruggiero LF (1994) American marten. In: Ruggiero LF, Aubry KB, Buskirk SW, Lyon LJ, Zielinski WJ, (ed) The scientific basis for conserving forest carnivores: American marten, fisher, lynx, and wolverine in the western United States. Gen. Tech. Rep. RM-254. Fort Collins, CO: US. Department of 10 Agriculture, Forest Service, Rocky Mountain Forest and Range Experiment Station. pp7-37 Clem MK (1977) Interspecific relationship of fisher and marten in Ontario during winter. In: Philips RL, Jonkel C (ed) Proceedings of the 1975 predator symposium, Missoula, MT, 16-19 June 1975. Montana Forest Conservation Experiment 15 Station, University of Montana, Missoula. pp165-182 Coffin KW, Kujala QJ, Douglass RJ, Irby LR (1997) Interactions among marten prey availability, vulnerability, and habitat structure. Martes: taxonomy, ecology, techniques, and management. The Provincial Museum of Alberta, Edmonton, Alberta, Canada, pp199–210 20 Corn JG, Raphael MG (1992) Habitat characteristics at marten subnivean access sites. J Wildlife Manage 56:442–448 Drew GS (1995) Winter habitat selection by American marten (Martes americana) in Newfoundland: why old growth? PhD thesis, Utah State University Krohn WB, Zielinski WJ, Boone RB (1997) Relations among fishers, snow, and martens 25 in California: results from small-scale spatial comparisons. Martes: taxonomy, ecology, techniques, and management. Provincial Museum of Alberta, Edmonton, Alberta, pp211–232 Potvin F, Belanger L, Lowell K (2000) Marten habitat selection in a clearcut boreal landscape. Conserv Biol 14:844-857 30 Raine RM (1987) Winter food habits and foraging behaviour of fishers (Martes pennanti) and martens (Martes americana) in southeastern Manitoba. Can J Zool 65:745–747 Robitaille JF, Aubry K (2000) Occurrence and activity of American martens Martes americana in relation to roads and other routes. Acta Theriol 45:137–143 Steele MA (1998) Tamiasciurus hudsonicus. Mammalian Species, American Society of 35 Mammalogists, pp1-9 Thompson ID, Colgan PW (1994) Marten activity in uncut and logged boreal forests in Ontario. J Wildl Manage 58:280–288 Wilson EBDE, Ruff S (1999) North American I Mammals. Smithsonian Institute, Washington, DC, USA 40 Zielinski WJ, Duncan NP. (2004) Diets of sympatric populations of American martens (Martes americana) and fishers (Martes pennanti) in California. J Mamm 85:470– 477 45 Supplement 2. Hypothesized cost of American marten (Martes americana) movement in 50 the upper peninsula of Michigan, USA in relation to marten harvest location and affiliation to one of three genetic clusters (indicated with different symbols). Cost surfaces represent a) presence of roads, b) canopy cover, c) percent forested area, d) percent coniferous forest, and e) fisher (Pekania pennanti) harvest density. Variables are described in Supplement 1. 55 Supplement 3. Results from sensitivity analysis to determine the influence of weighting scheme on the resulting cost distance from least cost path (LCP) analysis for each landscape feature. Model Geographic Distance (Euc) Roads (2 levels) (Rd) Canopy Cover (Can) Proportion Forested Area (For) Fisher Density (PP) Proportion Coniferous Area (Con) 60 Weights 1 1, 10 1,100 1, 1000 1 to 10 1 to 100 1 to 1000 1 to 10 1 to 100 1 to 1000 1 to 10 1 to 100 1 to 1000 1 to 10 1 to 100 1 to 1000 Mantel r 0.209 0.224 0.216 0.223 0.190 0.211 0.185 0.184 0.208 0.052 0.198 0.192 0.162 P value <0.002 <0.002 <0.002 <0.002 <0.002 <0.002 <0.002 <0.002 <0.002 0.035 <0.002 <0.002 <0.002 0.243 <0.002 0.221 <0.002 0.245 <0.002 Supplement 4: Description of Causal Modeling methods. We used causal modeling (Cushman et al. 2006; Cushman and Landguth 2010b) based on partial Mantel tests to quantify support (Mantel r) for each of our models and to inform our boundary analysis (described in Supplement 5) by identifying the landscape variables (given the set of 65 variables we quantified) that most highly correlated with genetic relatedness (Smouse et al. 1986; Cushman et al. 2006. We assessed statistical significance of each partial Mantel test at α = 0.0021 (Bonferonni correction of α = 0.05 based on multiple model comparisons). We conducted a series of partial Mantel tests between genetic distance and least cost distance corresponding to each of our landscape hypotheses, after partialing out 70 Euclidean distance (e.g., Genetic Distance ~ Cost Distance from Model 1 | Euclidean Distance; Cushman et al. 2006). We also conducted a series of partial Mantel tests between genetic distance and Euclidean distance, partialing out the least cost distance corresponding to each of our landscape hypotheses (e.g., Genetic Distance ~ Euclidean Distance | Cost Distance from Model 1). If cost distance estimated based on one or more 75 landscape features was significantly associated with genetic distance independent of Euclidean distance, we expected the former test to be statistically significant and the latter to be not significant (Cushman et al. 2006). Cushman SA, McKelvey KS, Hayden J, Schwartz MK (2006) Gene flow in complex landscapes: testing multiple hypotheses with causal modeling. Am Nat 168:486– 80 499 Cushman SA, Landguth EL (2010b) Spurious correlations and inference in landscape genetics. Mol Ecol 19:3592-3602 Smouse PE, Long JC, Sokal RR (1986) Regression and Correlation Extensions of the Mantel Test of Matrix Correspondence. Syst Zool 35:627-632 85 Supplement 5: Defining genetic clusters (additional details) The clustering algorithm implemented in program Geneland incorporates the spatial coordinates of the multilocus genotype for each individual to determine posterior probabilities of membership to a genetic cluster. We defined the boundaries of genetic 90 clusters as sharp gradients in posterior probabilities among different clusters. We assumed that the locational error associated with marten harvest location reporting was randomly distributed around the centroid of each township section (1 section = 2.6 km2), and included coordinate uncertainty of 1 km associated with marten harvest locations. We estimated the number of genetic clusters (K) using a Markov Chain Monte 95 Carlo (MCMC) algorithm. We first allowed K to vary and subsequently ran the algorithm with K fixed at the value most supported by the data (i.e., K from the initial run with the highest average posterior probability; Guillot et al. 2005b). We iterated this process four times with the following parameters: 250,000 MCMC iterations, maximum rate of Poisson process at 495 (i.e., equal to the number of individuals as suggested by Guillot et 100 al. 2005a), minimum K = 1, maximum K = 5, and maximum number of nuclei of the Poisson-Voronoi tessellation at approximately 3 times the maximum rate of the Poisson process (Guillot et al. 2005b). We used an uncorrelated allele frequency model (Guillot et al. 2005a). We calculated the mean logarithm of the posterior probability by re-running the MCMC algorithm 10 times (thinning = 10) with K fixed at 3 (previously inferred 105 number of genetic clusters when allowing K to vary). Guillot G, Estoup A, Mortier F, Cosson JF (2005b) A spatial statistical model for landscape genetics. Genetics 170:1261–1280 Supplement 6: Mantel and partial Mantel tests, comparing the relative influence of landcover variables in predicting American marten (Martes americana) gene flow. 110 Significant partial Mantel results are in bold. Mantel partial Mantel2 Model1 r P r P3 Con + Rd 0.226 <0.002 0.076 <0.002 Con 0.221 <0.002 0.056 <0.002 Can + Con 0.220 <0.002 0.055 <0.002 Rd 0.216 <0.002 0.037 <0.002 Can + Con + Rd 0.220 <0.002 0.056 0.004 Con + PP 0.219 <0.002 0.053 0.004 PP + Rd + Con 0.220 <0.002 0.052 0.004 Can + Con + PP + 0.218 <0.002 0.046 0.004 Rd Can + Con + PP 0.218 <0.002 0.045 0.006 PP 0.192 <0.002 0.028 0.076 Can 0.211 <0.002 -0.021 0.093 Can + PP 0.208 <0.002 -0.017 0.127 PP + Rd 0.201 <0.002 0.019 0.135 Can + Rd 0.210 <0.002 -0.017 0.158 PP + Rd + For 0.209 <0.002 -0.017 0.166 Can + For + Rd 0.209 <0.002 -0.017 0.171 PP + Rd + Can 0.209 <0.002 -0.015 0.176 Can + For 0.208 <0.002 -0.017 0.181 For + PP 0.197 <0.002 -0.017 0.211 Can + For + PP 0.209 <0.002 -0.010 0.292 Can + For + PP + 0.210 <0.002 -0.008 0.298 Rd For 0.208 <0.002 0.010 0.314 For + Rd 0.212 <0.002 0.007 0.323 Euc 0.209 <0.002 NA NA 1 Euc = Euclidean distance, Rd = Roads, Con = Proportion of coniferous forest, For= Proportion of area that is forested, PP = Fisher (Pekania pennanti) density, Can = Canopy cover. 2 115 Partial Mantel tests quantify the residual variation between each model and genetic distance, after accounting for variation associated with Euclidean distance (e.g., Can + Con | Euc). 3 Bonferroni correction of α = 0.05/24 = 0.0021). All univariate Mantel models correlating genetic relatedness to least cost paths were significant, with the highest Mantel r value corresponding to the model that included Con 120 + Rd (r = 0.226, P < 0.002; Supplement 6). After accounting for the variation associated with Euclidean distance (Euc) using partial Mantel tests, we found 4 significant models that described the boundaries of genetic relatedness (α < 0.0021): 1) Coniferous, 2) Roads 3) Canopy + Coniferous, and 4) Coniferous + Roads (Supplement 6). Additionally, the relationship between genetic relatedness and Euclidean distance was not significant after 125 partialing out the variation associated with each of the above 4 models (Supplement 7). The Coniferous + Roads model had the most support compared to all alternative models based on the magnitude of the partial Mantel r values (Supplement 6). Supplement 7. Partial Mantel tests comparing the relative influence of several landcover 130 variables in predicting American marten (Martes americana) gene flow. Significant partial Mantel results are in bold. partial Mantel P Euc | PP 0.10 <0.002 Euc | For + PP 0.09 <0.002 Euc | PP + Rd 0.08 <0.002 Euc | For 0.05 <0.002 Euc | Can + For 0.05 <0.002 Euc | PP + Rd + Can 0.05 <0.002 Euc | Can + For + PP 0.05 <0.002 Euc | Can + For + Rd 0.05 <0.002 Euc | Can + PP 0.05 0.003 Euc | Can + Rd 0.05 0.005 Euc | Can + For + PP + Rd 0.04 0.005 Euc | PP + Rd + For 0.05 0.006 Euc | Can 0.04 0.007 Euc | Rd -0.02 0.017 Euc | For + Rd 0.03 0.041 Euc | Con + PP 0.02 0.086 Euc | Can + Con + Rd -0.02 0.098 Euc | Can + Con -0.02 0.124 Euc | PP + Rd + Con 0.02 0.172 Euc | Can + Con + PP + Rd -0.02 0.197 Euc | Con + Rd -0.01 0.222 Euc | Can + Con + PP -0.01 0.237 Euc | Con -0.01 0.351 1 Partial Mantel tests quantify the residual variation between Euclidean distance and Model1 r genetic distance, after accounting for variation associated with each alternative landscape model. 135 2 Bonferroni correction of α = 0.05/24 = 0.0021)