Supplementary Information (docx 21K)
advertisement

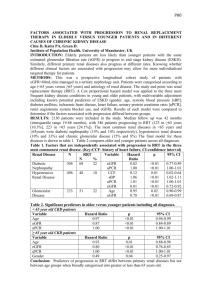
Supplementary Figure Legend Supplementary Figure 1. IGFBP2-induced STAT3 activation is mediated through EGFR. Immunoblot analysis of SNB19.BP2 cells depleted of EGFR by using 2 independent pools of EGFR siRNA (EGFR sir#1, EGFR sir#2) or scrambled negative control siRNA (ctrl siR), followed by overnight serum starvation and stimulation with 100ng/mL recombinant human IL6 (rhIL6) for 15 minutes. Supplementary Figure 2. Inhibition of ADAMs do not affect IGFBP2-mediated EGFR signaling activation. (A) U87 glioma cells were serum-starved overnight, then pretreated with an ADAMs inhibitor, TAPI-2 or marimastat (MMS) (20M), for 2 hours. After pretreatment, cells were stimulated with 100ng/mL IGFBP2 in serum-free medium with fresh TAPI-2 or marimastat (20M) for 5 minutes. Whole-cell lysates were collected for immunoblotting. (B) SNB19.EV and SNB19.BP2 cells were transfected with ADAM17 siRNA or scramble negative control siRNA for 48 hours and serum-starved overnight, then treated with 100ng/mL IGFBP2 for 5 minutes. Whole-cell lysates were collected for immunoblotting. Supplementary Figure 3. Hierarchical clustering of 157 experimentally validated STAT3 target genes from Ingenuity Pathway Analysis across all samples in the Rembrandt glioma dataset. Two distinct clusters formed and associated with tumor grade and IGFBP2 and STAT3 expression, but not with CTNN1B or FOXM1 expression. EGFR expression was elevated in a subset of glioblastomas. Blue bar represents low-grade glioma, and yellow bar represents high-grade glioma. 1 Supplementary Figure 4. Migration and invasion potential were significantly impaired in the SNB19.BP2ΔNLS cells. (A) Diagram of IGFBP2 domains and nuclear localization signal (NLS). (B) A migration assay was performed on SNB19.EV (empty vector), SNB19.BP2 WT (wild type) and SNB19.BP2 with a mutant NLS (SNB19.BP2ΔNLS or mutNLS) cells using a transwell migration chamber. (Left) Cells were fixed and stained after incubation for 4 hours. (Right) Bar graph represents the mean number of migrated cells in 5 random view fields (mean ± SD). (C) An invasion assay was performed on SNB19.EV, SNB19.BP2 WT and SNB19.BP2ΔNLS cells using a transwell invasion chamber. (Left) Cells were fixed and stained after incubation for 16 hours. (Right) Bar graph represents the mean number of invaded cells in 5 random view fields (mean ± SD). Indicated annotations correspond to the following P-values: *P<0.05, ***P<0.005, and ****P<0.0001. Supplementary Figure 5. IGFBP2 promotes nuclear EGFR accumulation in T98G glioma cells. (A) Immunoblot analysis comparing cytoplasmic and nuclear fractions of T98G cells depleted of IGFBP2 via 2 independent pools of IGFBP2 siRNA (BP2 siR #1, #2) for 48 hours to those treated with scrambled negative control siRNA (ctrl siR). Densitometric analysis of EGFR represented by the bar graph, demonstrates percentage of cytoplasmic (cyt) or nuclear (nuc) EGFR. (B) Immunoblot analysis of cytoplasmic and nuclear proteins in T98G cells transiently transfected with EV, BP2 WT and BP2ΔNLS. Densitometric analysis of EGFR represented by the bar graph, demonstrates percentage of cytoplasmic (cyt) or nuclear (nuc) EGFR. 2 Supplementary Figure 6. Mutation of the IGFBP2 NLS does not affect binding to EGFR. U87 glioma cells were transiently transfected with EV, BP2 WT or BP2ΔNLS plasmid followed by immunoprecipitation (IP) for IGFBP2 and immunoblotting (IB). Supplementary Table 1. Levels of IGFBP2, pSTAT3(Y705) and EGFR in a TMA of glioma samples from patients were evaluated using immunohistochemical analysis. Correlation levels were calculated by using the Pearson chi-square analysis. 3