How to write-up models with negative binomial variance Fred
advertisement

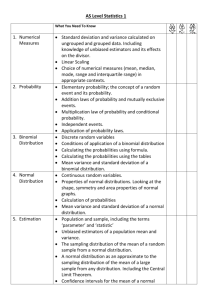
How to write-up models with negative binomial variance Fred Watson, 2010-2014 Negative binomial distributions are appropriate when your response variable is a positive integer with possibly large variance. Count data are typical of this. Conversely, if your response is a count, it is not appropriate to model it using continuous distributions (e.g. gamma, log-normal) or distributions that are defined below zero (e.g. normal). (Poisson distributions can also be used for count data, and are historically more common. Poisson models would be the best choice for equi-dispersed data, since they use one less parameter than negative binomial models, and are thus more parsimonious. You could choose between Poisson and negative binomial either using a dispersion test, or an AIC model comparison between two models that differ only in being either Poisson or negative binomial.) Let’s say you want to model the influence of weather on observed counts of a bird species. The response is the bird count, a positive integer. The predictor is weather, which we’ll say is a binary variable taking values of either 0 (clear weather) or 1 (cloudy weather). You could write: I modeled variation in bird count, Yi, at site i, as a function of weather, Wi, using a linear model with negative-binomial variance: 𝑌𝑖 ~ NB(𝜇𝑖 , 𝜃) 𝜇𝑖 = 𝛽𝐶 + 𝛽𝑊 𝑊𝑖 where NB(𝜇𝑖 , 𝜃)is a negative binomial distribution function with mean µi and a dispersion parameter (Venables & Ripley 2002), the mean count, µi, is modeled as linear function of weather Wi and C and W are fitted coefficients representing the mean count on clear days and the effect of cloudy weather on the count respectively. The model was fitted using the glm.nb function in the R statistical package (R Core Development team 2011) using the code: library("MASS") glm.nb( Y~W, link="identity") It’s probably more common to use a log link function, which leads to an exponential relationship between mu and the predictor variable/s. So if you did that, you’d need to change the equation, code, and text as indicated in red below: I modeled variation in bird count, Yi, at site i, as a function of weather, Wi, using a linear model with negative-binomial variance: 𝑌𝑖 ~ NB(𝜇𝑖 , 𝜃) 𝜇𝑖 = exp(𝛽𝐶 + 𝛽𝑊 𝑊𝑖 ) where NB(𝜇𝑖 , 𝜃)is a negative binomial distribution function with mean µi and a dispersion parameter (Venables & Ripley 2002), the mean count, µi, is modeled as an exponential function of weather Wi and C and W are fitted coefficients representing the mean count on clear days and the effect of cloudy weather on the count respectively. The model was fitted using the glm.nb function in the R statistical package (R Core Development team 2011) using the code: library("MASS") glm.nb( Y~W, link="log") A more-complex way to write this might be: I modeled variation in bird count, Yi, at site i, as a function of weather, Wi, using a linear model with negative-binomial variance: 𝑓𝑌𝑖 (𝑦|𝜇𝑖 , 𝜃) = NB(𝑦|𝜇𝑖 , 𝜃) = Γ(𝜃 + 𝑦) 𝜇𝑖 𝑦 𝜃 𝜃 Γ(𝜃)𝑦! (𝜇𝑖 + 𝜃)𝜃+𝑦 𝜇𝑖 = 𝛽𝐶 + 𝛽𝑊 𝑊𝑖 where µi is a linear model for the mean count, C and W are fitted coefficients representing the mean count on clear days, and the effect of cloudy weather on the count, 𝑓𝑌𝑖 (𝑦|𝜇𝑖 , 𝜃) is the probability mass function for Yi, is a dispersion parameter, and (.) is the gamma function. This model leads to: E(𝑌𝑖 ) = 𝜇𝑖 var(𝑌𝑖 ) = 𝜇𝑖 + 𝜇𝑖 2 /𝜃 where E(Yi) denotes the expected value of Yi, and var(Yi) denotes the variance of Yi, from which it can be seen that the variance increases with the mean and can be larger than the mean (Venables & Ripley 2002). The model was fitted using the glm.nb function in the R statistical package (R Core Development team 2011) using the code: library("MASS") glm.nb( Y~W, link="identity")