SUPPLEMENTARY MATERIALS AND METHODS Human Tumor
advertisement

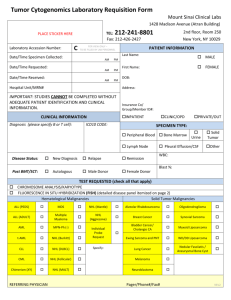
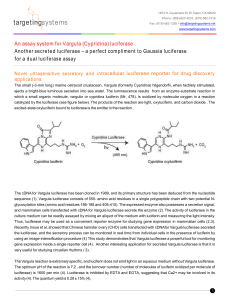
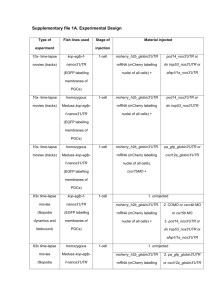
SUPPLEMENTARY MATERIALS AND METHODS Human Tumor Samples For this IRB approved study, tissues were obtained from patients undergoing surgical resection and who were consented to the institution’s tissue collection protocol at Moffitt Cancer Center between 2002-2012. Histopathological examination of specimens (histological type, tumor size etc.) was performed by an experienced GI pathologist (B.C.). Macrodissection of formalin-fixed paraffin-embedded (FFPE) tissues was performed to isolate tumor sections (containing >80% tumor cells) for RNA extraction. Microarray RNA was extracted from FFPE material using the RecoverAllTM Total Nucleic Acid Isolation kit optimized for FFPE samples (Ambion, Austin, TX) according to the manufacturer’s protocol. All RNAs were quantified by using the Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Pittsburgh, PA). Microarray analysis was performed using the oligonucleotide probe-based Affymetrix U133 plus 2.0 chips per standard Affymetrix protocols. Biotin-labeled cRNA generated by in vitro transcription was fragmented and hybridized to the gene chip arrays at 45ºC for 16 hr and then washed and stained with gene chip fluidics according to Affymetrix protocol. The probe arrays were then scanned at 1.5-um resolution using the Affymetrix GeneChipScanner 3000. Array Data Analysis Scanned output files were visually inspected for hybridization artifacts and then analyzed by using Affymetrix Microarray 5.0 software. Arrays were scaled to an average intensity of 500 and analyzed independently. The MAS 5.0 software used for statistical algorithm to determine the signal intensity of a transcript from the behavior of 11 different oligonucleotide probes designed to detect the same gene. Probe sets that yield a change pvalue less than 0.006 was identified as changed. With replicate measurements the p-value was adjusted. Alternatively, the signal intensity was calculated by the robust multi-array analysis method (RMA) developed by Irizarry et al. Gene changes were then selected using the fold-change cut-off available in RMA or using the Significance Analysis of Microarrays (SAM) technique. miRNA quantification Total RNA was isolated from macrodissected FFPE tissues using the mirVANA™ miRNA Isolation Kit (Ambion, Austin, TX) according to the manufacturer’s protocol, including enrichment for small RNAs. RNA concentration and quality were determined using NanoDrop1000. Only RNA samples of sufficient quality were used. Serum AFP quantification Patient serum and growth media from cell lines were utilized for quantification of AFP in the clinical chemistry laboratory at the Moffitt Cancer Center. Tumors were considered to be AFP-secreting if the patient’s serum AFP level was above the upper limit of normal (37IU) reported by our laboratory. Quantitative RT –PCR Analysis The differential expression level of mir-675 in HCC tumor samples and hepatoma cell lines was confirmed by quantitative real-time PCR analysis. Total RNA was extracted from tumor tissues and hepatoma cells. 1 ug of total RNA was reverse transcribed using TaqMan reverse transcription reagent (Applied Biosystems, Carlsbad, CA). cDNA was amplified in a 9700 Gene Amp PCR System (Applied Biosystems, Carlsbad, CA). Using specific primers and probe for mir-675 (NCBI reference sequence NR_030533.1) (Forward: 5’- AGG GTC TGG TGC GGA GA -3’ Reverse: 5’ GAG CGG TGA GGG CAT ACA-3’ Probe: 5’-TCA CCA AGT CCA CTG TGG GC 3’). Experiments were conducted in triplicate. Amplification reactions were run using 7900 HT real-time PCR instrument (Applied Biosystems, Carlsbad, CA). Results were reported as the ratio between the expression of mir-675 gene and the expression of Glyceraldehydes-3phosphate dehydrogenase. Biological Pathway Analysis MetaCore pathway analysis by GeneGo was used to identify the Gene GO pathways and Gene GO networks for genes with statistically significant changes in expression, when comparing transcriptomes of AFP-secreting and non-secreting human tumor samples as well as miR-675 overexpressing and control hepatoma cells. Significance was calculated using a built-in function of MetaCore software that utilizes a variation of the Fisher's exact test adjusted for multiple sample testing with the Benjamini-Hochberg FDR analysis. MicroRNA Target Prediction In silico microRNA miRNAMap 2.0 target prediction software was utilized to identify potential targets of miR-675 (http://miRNAMap.mbc.nctu.edu.tw/). The DNA sequences of the 3’-untranslated region (UTR) region of Twist1 and Rb mRNA was obtained from Genbank of the National Center for Biotechnology Information webpage (http://www.ncbi.nlm.nih.gov/). Cell Culture and Transfection The HepG2, SK-Hep and Hep3B cell lines were obtained from the ATCC. The cell lines were authenticated in January 2012 by short tandem repeat (STR) analysis at the Genetic Resources CoreFacility ay Johns Hopkins University using an Identifiler kit and GeneMapper software (Applied Biosystems, Carlsbad, CA). Transfections were performed in 6 well plates using Lipofectamine-2000 (Invitrogen, Carlsbad, CA) and 50 ng/2ml of pre-miR-675, negative control miRNA, anti-miR-675 or anti-miR control (Applied Biosystems, Carlsbad, CA) according to the manufacturer’s protocol. Proliferation 1 x 103 cell/ well were seeded in quadruplicate in 96-well plates. Cells were incubated for 72 hr, and cell proliferation was assayed using the XTT Cell Proliferation Kit II (Roche, Mannheim, Germany). Proliferation was scored by measuring the spectrophotometrical absorbance (wave length 540 nM) of the samples. Cell Cycle Distribution assay Analysis of cellular discrimination in Go/G1 versus S versus G2/M phases of the cell cycle was conducted by measuring the cellular DNA content with propidium iodide methodology. One million cells were washed in 1X PBS and transferred into tube containing 70% ethanol. The cells were kept overnight at -20°C and fixed in 70% ethanol and then rinsed with 1X PBS. The cells were then suspended in 1ml PI/Triton solution in the presence RNase A. The cells were kept for 15 minutes at 37°C incubator. The cell fluorescence was measured in the flow cytometer. Western blot Analysis Cell lysis and protein extraction were achieved using the TRIZOLkit (Invitrogen, Carlsbad, CA). Extracted proteins were dissolved in 1% SDS and concentrations were measured using the Bio-Rad protein assay kit (Bio-Rad). Protein (50 to 100 ug) per sample was loaded and fractioned by electrophoresis on SDS-PAGE gel. Proteins were then transferred to a nitrocellulose membrane using Trans-Blot SD, Semi -Dry Transfer Cell (Bio-Rad, Hercules, CA). The membranes were blocked with 5% non-fat dry milk for 2 hr and washed with TBST solution. Primary antibodies (1:1000) were diluted into a dilution buffer, comprised of 1X TBS, 0.01% Tween-20 with either 5% BSA or 5% nonfat dry milk. The membranes were hybridized at 4ºC overnight and then washed. Membranes were then reacted with secondary antibodies (1:2000) for two hours, washed and developed using a chemiluminescent kit (commercially available antibodies: Amersham, Piscataway, NJ, Cell Signaling, Beverly, MA; Santa Cruz Biotechnology, Santa Cruz, CA). Soft Agar Assay To make the bottom layer, a 3% Bacto agar (BD Biosciences, San Jose, CA) stock was made in DIDW and sterilized in the autoclave. At 48°C, 40 mL of the growth medium were mixed with 10 mL of the 3% agar. 2mL/well was plated into the wells of a 12-well tissue culture plate and solidified at room temperature. For the cell layer, the cells were counted using trypan blue dye and a hematocytometer. 3000 cells/well were prepared and resuspended in separate tubes, in which agar medium were mixed with the cells. Oneand-a-half mL of cell-agar mixture were dispensed on top of the base layer. The plates were then incubated for 3 weeks. Cell line used in triplicates for the analysis. In order to develop the plates, 1mg/mL MTT was dissolved in the growth medium. 150ul of MTT dye was added in each well and incubated overnight. The colonies were counted and photographed at microscopic core facility. Invasion Invasive potential was measured using CytoSelect 24-well Cell Invasion Assay (Cell Biolabs Inc, San Diego, CA). Cells that invaded the lower surface of the membrane were stained with DAPI (Invitrogen, Carlsbad, CA) and counted per 100 high-power fields with an inverted microscope. All experiments were run in duplicate and performed on two separate occasions. Luciferase Reporter Assay The 3’UTR clone of Twist1 (NM_000474) was purchased from Origene, Rockville, MD. The pmiR target vector containing Twist1 3’UTR (Origene, Rockville, MD) was transformed into competent cells (DH5α) and grown. Plasmid DNA was extracted and measured. The restriction digest was performed with SgfI and MluI as recommended by the manufacturer and confirmed the product size of Twist1 3’UTR. The plasmid DNA with Twist1 3’UTR was sequenced and validated with gene bank sequence. Pre-miR-675 and the Negative Control (Pre-miR) were purchased from Ambion, Austin, TX. A total 250K human HepG2 cells were seeded in each well of a 6 well plate; as the cells became 60-70% confluent, the transfection assay was performed. There are 3 triplicates for control and 3 triplicates for case. 1000ng of plasmid DNA and 50nM of pre-miR or control miRNA were co-transfected into HepG2 cells for 48 hours. Each sample was also cotransfected with 0.5ug of pRL-CMV plasmid-expressing Renilla luciferase to monitor the transfection efficiency. At 48h post transfection, the activity of firefly luciferase was measured by using the dual-luciferase reporter assay system as described by the manufacturer (Promega, San Louis Obispo, CA). Relative luciferase activity was normalized with Renilla luciferase activity. Statistical Analyses Determination of biological pathway dependence was undertaken using the basic formula for hypergeometric distribution, with consideration given to the number of differentially regulated genes, the number of genes in a given pathway, and the number of genes in all pathways. Results of individual experiments were analyzed using 2-sided Student's t-test with significance accepted with > 95% confidence. For microarray analysis, refer to section 2.3.