tpj12722-sup-0010

Supporting Information Legends
Table S1.
List of up-regulated genes in shoot at pH 5.8 classified in the Venn diagram shown in
Figure S4.
Table S2.
List of down-regulated genes in shoot at pH 5.8 classified in the Venn diagram shown in
Figure S4.
Table S3.
List of up-regulated genes in root at pH 5.8 classified in the Venn diagram shown in Figure
S4.
Table S4.
List of down-regulated genes in root at pH 5.8 classified in the Venn diagram shown in
Figure S4.
Table S5.
List of up-regulated genes in shoot at pH 8.0 classified in the Venn diagram shown in
Figure S5.
Table S6.
List of down-regulated genes in shoot at pH 8.0 classified in the Venn diagram shown in
Figure S5.
Table S7.
List of up-regulated genes in root at pH 8.0 classified in the Venn diagram shown in Figure
S5.
Table S8.
List of down-regulated genes in root at pH 8.0 classified in the Venn diagram shown in
Figure S5.
Table S9.
Primers used in this study.
Table S10.
Gene names and codes shown in Supplemental Figure S3.
Figure S1. Effects of EDTA application on rice seedling growth at pH 8.0. One-week-old rice seedlings germinated and grown hydroponically in deionized water were transferred to nutrient solution containing 0, 0.3, 3, 30, or 150 µM EDTA at pH 8.0, and further cultivated for 2 weeks.
SPAD value was measured using a chlorophyll meter (SPAD-502). All treatments contained equal
amounts of iron. Average values and standard deviation were calculated with five biological replicates.
Asterisks indicate significant difference from control (0 µM EDTA) at pH 8.0 (**, p < 0.01, Student’s t -test).
Figure S2.
Iron distribution in rice seedlings treated with DMA or EDTA for 22 h. Iron transportation from root to shoot was visualized using 55 Fe with the BAS-5000 system. (a) and (b), autoradiograph of 55 Fe with different chelators under normal- and high-pH conditions (pH 5.8 and 8.0), respectively.
(c) and (d), photograph of rice seedlings used for autoradiograph analysis shown in panel (a) and (b), respectively. Scale bar = 4 cm. Control, no chelator in the medium. All treatments contained equal amounts of iron. Values shown are mean ± SD ( n = 5).
Figure S3. Expression profile of iron and nitrate (nitrogen) assimilation related genes. Microarray analysis was performed using RNA extracted from shoots and roots of seedlings in indicated treatments at pH 5.8 and 8.0. Genes in each compartment are listed in supplemental Tables S1 to S8.
Heatmap was generated using heatmap.2 program in R with log ratio values normalized to the median by GeneSpring. Gene codes for representative genes are listed in Supplemental Table S10.
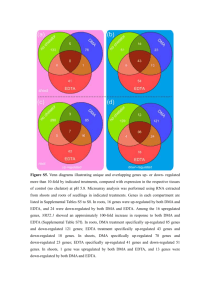
Figure S4. Venn diagrams illustrating unique and overlapping genes showing more than 10-times up- or down-regulated expression compared with that in the no-chelator control at pH 5.8. Microarray analysis was performed using RNA extracted from shoots and roots of seedlings in indicated treatments at pH 5.8. Genes in each compartment are listed in Supplemental Tables S1 to S4. In roots,
19 genes were up-regulated and 88 were down-regulated in response to both DMA and EDTA.
Among the 19 up-regulated genes was NRT2.1
, which showed an approximately 80-fold increase in roots in response to both DMA and EDTA (Supplemental Table S3B). In roots, DMA specifically upregulated 34 genes and down-regulated 35 genes, while EDTA specifically up-regulated 47 genes and down-regulated 21 genes. In shoots, DMA specifically up-regulated 55 genes and down-regulated 24 genes, while EDTA specifically up-regulated 57 genes and down-regulated 17 genes. In shoots, no genes were upregulated by both DMA and EDTA, while 90 genes were down-regulated by both DMA and EDTA.
Figure S5. Venn diagrams illustrating unique and overlapping genes up- or down- regulated more than 10-fold by indicated treatments, compared with expression in the respective tissues of control (no chelator) at pH 5.8. Microarray analysis was performed using RNA extracted from shoots and roots of seedlings in indicated treatments. Genes in each compartment are listed in Supplemental Tables S5 to
S8. In roots, 16 genes were up-regulated by both DMA and EDTA, and 24 were down-regulated by
both DMA and EDTA. Among the 16 upregulated genes, NRT2.1
showed an approximately 100-fold increase in response to both DMA and EDTA (Supplemental Table S7f). In roots, DMA treatment specifically up-regulated 85 genes and down-regulated 121 genes; EDTA treatment specifically upregulated 43 genes and down-regulated 18 genes. In shoots, DMA specifically up-regulated 78 genes and down-regulated 23 genes; EDTA specifically up-regulated 41 genes and down-regulated 51 genes.
In shoots, 1 gene was upregulated by both DMA and EDTA, and 13 genes were down-regulated by both DMA and EDTA.
Figure S6.
Transcription analysis to validate microarray analysis of gene transcription in roots and shoots. Experimental design was the same as that used to generate data shown in Figure 6. Black bars, shoots; white bars, roots. Asterisks indicate significant difference from control (no chelator) at pH 5.8 or 8.0 (*, p < 0.05; **, p < 0.01, Student’s t -test). Values shown are mean ± SD ( n = 3).
Figure S7. Effect of chelator treatments on enzyme activity of NADH- and NADPH-dependent nitrate reductases in roots of rice seedlings. DMA or EDTA was added to medium at a final concentration of 30 µM. Control, no chelator in the medium. Assays were separately performed to measure NADH- and NADPH-dependent nitrate reductase activities. Equal amounts of iron were present in all treatments. Asterisks indicate significant difference from control (no chelator) at pH 5.8 or pH 8.0 (**, p < 0.01; Student’s t -test). Values shown are mean ± SD ( n = 3).
Appendix 1. Additional experimental procedure: microarray analysis.