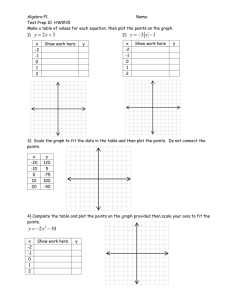
Exercise #2
advertisement

Weizmann 2013-2014 Introduction to Matlab & Data Analysis Exercise #3 - Simple Data Analysis and Graphics Tutor in charge of this HW: Gil Farkash, e-mail for Questions: gil.farkash@weizmann.ac.il Last day for submission: XXX The mail subject should be: 'matlab intro exercise 3'. Attach only the script file to your mail. HW instructions: Please name your script "hw3_<ID 1>_<ID 2>.m". Do not use loops in this ex. Follow the HW guidelines carefully. Please pay attention to the figure requirements (title, axis labels, subplots…) All numbers should be assigned to variables Data description: Download the file MSB_2005.mat. This file contains the expression data of 170 genes in 12 duplicate time points (totally 24 samples) from Tabach et al MSB 2005. Briefly, the values in the file are gene expression measurements reflecting the number of mRNA molecules of each gene. These values were obtained using a high throughput method called microarray that simultaneously measures the expression values of multiple genes. The raw data was further processed and normalized to filter noises from the expression signal. Different columns describe different types of cells in different cancer development stages. Motivation: In this HW you will interrogate the data using simple analysis and visualization functions. You will learn how to visualize a large set of genes, which is a routine task in many biological and bioinformatics studies. You'll find attached to this ex an Excel file containing the same data as in the MSB_2005.mat file. This spread sheet should help in understanding the dataset. You can use the Excel file to understand the origin of the variables “genes”, “samples” and “data”. 1 Weizmann 2013-2014 Introduction to Matlab & Data Analysis Question 1- the plot Load the file MSB_2005.mat. You should have 3 variables – 1. “genes” – names of the 170 genes in this data set. 2. “samples” – 12 samples in duplicates, total of 24 samples. 3. “data” – a 170x24 matrix. Each row is a gene measured in the 24 samples. Calculate the mean and the median expression of the 170 genes along the different samples (along the columns) and store the information in the appropriate variables. Use short and meaningful names for your new variables. You can use the help command to learn about the mean and median functions. Read about the function “trimmean”. Calculate the data “trimmean” using 20%, and store it in an appropriate variable. Answer in your script: when is it better to use “trimmean” or “median” over the regular mean calculation? Figure 1: First, we will visualize the behavior of the genes average, median and trimmean in each sample. Plot the mean, median and trimmean variables in the same graph, each one with a different color. The mean plot should be red stars connected by a solid line, the median should be blue circles connected by a dotted line, and the trimmean should be green x-marks connected by dashed line. Line width should be 2. Add the title "mean median and trimmean expression of 170 genes". The x label should be "Samples" and y label should be "Expression". Add legend in the lower right corner of the axes, so it won’t cover the data lines. (Hint: you can use the functions: title, legend, xlabel, ylabel, plot and hold on). Figure 2: Create a scatter plot of the mean versus the median for each sample (see help). Calculate the minimal and maximal values among both datasets. Add to the maximal value 0.1, decrease from the minimal value 0.1. Plot in the same axis the x=y linear line between the new minimal and maximal values. This line should be a red solid line. Set the axis limits to be the minimal and the maximal values that you found and calculated. Add the title "mean versus median expression of 170 genes". The axis labels should be “mean” and “median” corresponds to the data presented in the figure. Figure 3: Plot again Figure 1 this time use the bar function instead of “plot” in order to create Figure 1. Don’t forget to add axis labels, title and legend in the lower right corner. 2 Weizmann 2013-2014 Introduction to Matlab & Data Analysis Figure 4: Plot the expression values of genes number 3, 28, 53, 78 , 103 etc… (one in every 25 genes, stars from 3) in a single plot. Each gene should be represented as a connected line with a different color. (Reminder: This can be done by giving the plot function a matrix). Add axis labels as in the previous figs, and the title “Expression data of 7 genes”. Add an appropriate legend in the lower right corner. Find the gene that had the maximal expression value in the entire data. Find the gene that had the minimal expression value in the entire data. Find the gene that changed the most over all conditions, i.e. the gene that has the largest absolute difference between its first value and its last value. Figure 5: Plot the expression values of the “max gene”, the “min gene” and the “max changed gene”, add axis labels and a legend. The legend format should be: max gene: <the gene name> min gene: <the gene name> max changed gene: <the gene name> Add axis labels as in the previous figs, and the title: “The min max and max changed genes expression data” The first three graphs should be in the same figure and should be arranged like this (the first two graphs in the upper row, and the third on the second row): 3 Weizmann 2013-2014 Introduction to Matlab & Data Analysis The other two figures should be on the same figure, and should look like this: Please make sure your results look the same as the figures attached. Question 2 In this question we will visualize the expression data using the function imagesc (for details on this function use help). Figure 1: Create a new variable that contains the average of the duplicates (sample 12, 3-4, 5-6 ……23-24 are the duplicates). This variable should have the size of 170 rows and 12 columns. Use the imagesc function to plot it. Don’t forget to add a title and axis labels (“Samples”, “Genes”). Use the function colorbar to include the color bar on the right side of the figure. Remove the “y-axis” ticks. Figure 2: Use the function colormap (redgreencmap) to create a red green version of the plot. What is the approximate range of the values in each sample? Figure 3: Create a figure using the function caxis([-0.5 0.5]) (see help) . What happened to the colors? What will happen to the colors if you change the numbers from 0.5 and -0.5 to 1 and -1, or -0.2 and 0.2? In which cases will you use this function? Your script should not create any figure in this question, (you should 4 Weizmann 2013-2014 Introduction to Matlab & Data Analysis create the figures in order to answer the theoretical question but do not include them in your script), the script should contain the answers to the theoretical questions. Figure 4: Choose your favorite colors and use the colormap to create new a colorful image. (Hint: black is not colorful) Your plots should look like this: (2.1 in a separate figure, 2.2 and 2.3 – two subplots in the same figure side by side, and 2.5 in a separate figure). 5 Weizmann 2013-2014 Introduction to Matlab & Data Analysis And another plot with your favorite colors. You should have 2 figures from question1, and 3 figures in the second question, total of 5 figures. 6