Cloning_protospacers_into_PX_vectors_7_23_14
advertisement

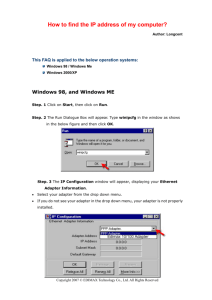
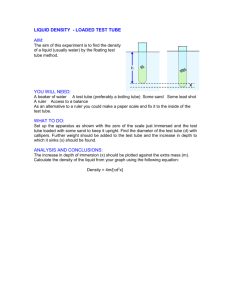
Cloning protospacer adapters into PX330/PX335/PX458/PX459 (Zhang lab, MIT) via BbsI site. I. Anneal top and bottom oligos of adapter 1. Resuspend oligos in water to 1 µg/µl concentration. 2. Combine the following in a 200 µl PCR tube: a. 5 µl “top” oligo b. 5 µl “bottom” oligo c. 10 µl of 10x annealing buffer d. 80 µl water (10X Annealing Buffer: 1 M NaCl / 10 mM EDTA pH 8.0 / 100 mM Tris pH 7.5) 3. Mix well. Run the mix in a thermal cycler with an annealing program, such as: Step (1) 94˚ for 3 minutes; Step (2) cool from 94 ˚ to 25˚ slowly, such as over a 30 minute period. Alternatively to a thermal cyler: Heat a beaker of water to boiling. Float the tube in the boiling water bath for 5’. Remove the beaker from the heat and let it cool off naturally on a benchtop, till it is room temperature. 4. Transfer the annealed adapter to a 1.5 ml tube. Add 900 µl water. The final concentration of annealed adapter is now ~ 10 ng/µl. II. Ligate to BbsI-cut vector. 1. Before you start: You will need to have the vector DNA previously cut with BbsI and gel-purifed, and in 10 mM Tris or Lo-TE at a concentration of at least 10 ng/µl. 2. I use the NEB Quick Ligation kit, and I reduce the volumes by half to save reagents. It works great. Combine the following in this order: a. 2.5 µl of 10 ng/µl BbsI-cut vector b. 1 µl of 10 ng/µl annealed adapter c. 1.5 µl of water d. 5 µl of 2x Quick Ligase buffer e. 0.5 µl of Quick Ligase enzyme 3. Mix briefly, and incubate at room temp. for 5’. Use immediately for transformation. III. Transform DH5 cells. 1. You will need a 42˚ water bath and LB+AMP plates. You will also need to get a tube of competent DH5 cells from the 9th floor core in Light Hall. Thaw the cells on ice. 2. Transfer 100 µl cells to a 1.5 ml tube. 3. Add 5 µl of the ligation reaction. Mix well (do not vortex). 4. Incubate 45’ on ice. 5. Heat-shock the cells at 42˚ for 2’. 6. Transfer cells back to ice. 7. Add 900 µl of LB media to the cells. Transfer the cells to a 15 ml tube. 8. Recover the cells by incubating in a 37˚ shaker incubator for 30’ at 250 rpm. 9. Plate out 100 µl of the cells on an LB+AMP plate. Incubate at 37˚ overnight. You should get at least a few dozen colonies (e.g. 10-100 is typical). Most of 1 them will contain the correctly cloned product. When I have done negative controls with no adapter, I get zero or just a couple of colonies. IV. Screen colonies for correct insertion of adapter. 1. Inoculate 2 colonies separately into 15 ml tubes with 2 mls of LB+100 µg/ml AMP. Shake overnight at 37˚. 2. Perform Qiagen minipreps on 1.5 ml of each plasmid culture. Save the remaining ~0.5 ml for making a glycerol stock to store at -80˚. (see below). The culture sample can be stored at 4˚ for a few days before you make the glycerol stock. 3. Spec the DNA. Usually the concentration is about 40-100 ng/ul. Prepare a sample for direct Sanger sequencing using this primer: U6F1: TACGATACAAGGCTGTTAGAGAG This sequencing will read-through the region of the BbsI site and verify that the adapter has inserted properly. V. Make glycerol stocks of the correct clones. To the remaining ~0.5 ml of the miniprep culture in step IV above, add 0.5 ml of sterile 30% glycerol. (Glycerol solutions should be sterilized by filtration, never autoclaving.) Mix and store the glycerol stock at -80˚. Doug Mortlock 7/23/14 2