Thibodeau, Walsh and Beisner - Supplementary Material Appendix
advertisement

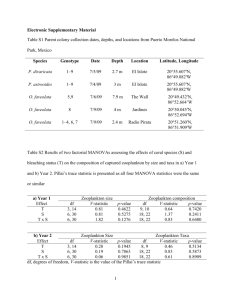
1 Thibodeau, Walsh and Beisner - Supplementary Material Appendix A: Zooplankton responses in the mesocosm experiment Zooplankton communities were established to minimize variation between mesocosms. However, we did sample their communities as well to determine whether there was important treatment variation that could explain the variation in phytoplankton community responses observed. Zooplankton were sampled on June 5, June 26 and July 10, 2012 via vertical net hauls from 3m to the surface using a 30µm mesh net (20cm diameter, 59cm long). Animals were anesthetized prior to preservation in 70% ethanol. In the lab, crustacean zooplankton were identified and counted using an Olympus dissecting microscope. Zooplankton responses to perturbation To assess zooplankton community response to perturbation and consequently to infer top-down effects on phytoplankton, we performed four two-way repeated measures ANOVAs (using the Anova function of the Car package in R). Factors were the treatments (Control, Pulse, Press) and time, with three levels corresponding to the dates on which we enumerated zooplankton abundances: June 5th (PRE-acidification, day 157), June 26th (POST 1, day 178) and July 10th (POST 2, day 192). We tested for effects within (i) the whole community with all zooplankton taxa included, (ii) only copepods (total, adults only and nauplii only), (iii) and all pelagic cladocerans (i.e. no chydorids), and (iv) chydorids only. Chydorids were analysed separately from other cladocerans as they have a particular feeding strategy adapted to littoral environment, crawling on and scraping food from surfaces (Dodson and Frey 2001), including the surface of mesocosm bags. Where an interaction between the two factors was significant, we performed a simple ANOVA and a Tukey-HSD test separately on all levels of the significant 2 factor in a one-way repeated measures ANOVA to determine which group at which time point or in which treatment was different from the others. The two-way repeated measures ANOVA showed several significant interactions between treatment and time for all four levels of taxonomic grouping (Table A1 for means by date and treatment). For total community abundances (interaction, p=0.006; two-way ANOVA), perturbation treatment type was the significant factor in the one-way repeated measures ANOVA with a significantly lower abundance of zooplankton in the Press mesocosms on POST1, three weeks after perturbation (p=0.007; one way ANOVA), but nor for the PRE or POST2 dates. There was also an interaction between time and treatments for the copepods (p=0.0007; two-way ANOVA). One-way ANOVAs showed that copepod abundances were significantly lower in the Press treatments than in either the Pulse or the Control on both the post-acidification dates. Additionally, there was a significant abundance peak in copepod abundance in the Pulse mesocosms on the POST1 date (p=0.005; one-way ANOVA). ANOVAs separating more advanced stages (copepodites and adults) from neonate (nauplii) copepods showed that this peak was a result of an increase in nauplii (p=0.006; one way ANOVA). For the advanced stages, there was no significant difference between either treatment or time points. For pelagic cladocerans (interaction p=0.025; two-way ANOVA), abundances were significantly less on both post-perturbation dates in acidified treatments. Finally, interaction between factors in the chydorids analysis was marginally significant (p=0.0497; two-way ANOVA). The simple ANOVAs showed that there were significantly more chydorids in the Press mesocosms on the first post-acidification date (POST1) (p=0.025; one-way ANOVA). 3 Zooplankton influence on phytoplankton community biovolume responses We explored the relationship between phytoplankton community biovolume (biomass) and pelagic zooplankton abundance (pelagic cladocera and copepods) with treatment as a covariable in ANCOVA in R using the three POST1 and three POST2 observations in each treatment. The goal was to test whether pelagic zooplankton abundance influenced the response of phytoplankton biomass to the pH Treatments (Pulse, Press and Control). The interaction between Treatment and zooplankton abundance was non-significant (P=0.251) indicating no significant difference between slopes in the phytoplankton biomass response. Discussion Given that we wanted to assess the phytoplankton community response to acid perturbation in a way that resembled natural lake conditions as much as possible, we included zooplankton in the experimental mesocosms. As would be expected, zooplankton community structure was also affected to some degree by acidification and this may have impacted in part the phytoplankton community response; although it appears to not have directly affected the size of the total biovolume increases following acidification. With respect to changes in zooplankton composition, peaks in chydorids and nauplii on the POST1 date (day 178) in the Press and Pulse treatments respectively would have had little, to no impact on focal pelagic phytoplankton responses as chydorids graze periphyton growing on the mesocosm walls and nauplii feed preferentially on smaller protozoa than on our response species (Turner 2004). The main pelagic zooplankton response consisted of reduced copepod biomass in the Press and reduced pelagic cladocera biomass post-perturbation in both acidified treatments. Such herbivore declines would have relieved predation pressure on the focal phytoplankton community and could have enabled the phytoplankton compensatory dynamics 4 observed via growth stimulation, thereby representing an indirect effect of acidification on phytoplankton. Thus, this effect through zooplankton still represents an overall perturbation effect because this decrease in important zooplankton grazers was part of the overall plankton community response to acidification. Our study thus provides information on how a natural plankton community would react to a sudden acidification, informing us about the cumulative ecological impact of both direct and indirect forces of acidification on plankton communities. With respect to the genetic response to perturbation by phytoplankton however, we believe that its source was a direct result of the perturbation and not from predation release. Both drift and evolutionary rescue have been shown to occur after significant declines in abundances, which would not result when predatory pressure is relieved (i.e. loss of zooplankton owing to acidification that we observed), but rather would occur as a result of the perturbation alone. The genetic shifts we observed in phytoplankton could thus only be a result of acidification. Predation release, on the other hand, would instead liberate phytoplankton from selective predation pressure, thereby not inducing a rapid evolution to adapt to a new predatorfree condition, as would a drastically lowered pH. Consequently, while liberation from zooplankton predation might be involved in the morphospecies responses of phytoplankton communities, it is unlikely to affect its genetic response to acidification. References Turner, M., E. Howel, M. Summerby, R. Hesslein, D. Findlay, and M. Jackson. 1991. Changes in epilithon and epiphyton associated with experimental acidification of a lake to pH 5.0. Limnology and Oceanography 36: 1390–1405. 5 Table A1: Zooplankton mean±s.e. biomass in µg/L, by treatment and time for each of the main taxonomic groupings. Taxonomic group PRE (157) POST1 (178) Control Pulse Press Control Whole 45.2±21. 52.9±7.6 66.0±11.6 54.1±20.1 community 3 Copepods 7.5±2.4 13.0±7.8 14.7±6.4 Cladocera Pulse POST2 (192) Press Control Pulse Press 61.4±12.2 9.3±3.6 29.9±14.9 19.8±9.3 5.1±2.0 26.4±15.5 48.3±8.2 0.7±0.8 18.8±9.2 19.6±9.5 0.4±0.6 35.4±22.7 39.9±1.2 51.3±10.8 27.0±23.0 12.4±9.4 0.8±0.9 10.7±9.2 0.1±0.1 2.9±2.5 Chydorids 2.3±3.7 0.05±0.08 0 ±0 0.6±0.7 0.7±0.5 7.9±4.6 0.3±0.1 0.1±0.1 1.8±1.4 Copepods 0.4±0.3 0.6±0.6 2.4±3.8 12.2±8.1 2.8±1.7 0.4±0.4 1.4±0.7 1.7±2.0 0.4±0.5 7.1±2.3 12.4±7.3 12.3±2.6 14.2±7.5 45.5±9.2 0.2±0.3 17.5±9.5 17.8±8.1 0.05±0.08 (pelagic) (mature) Copepod nauplii 6 Appendix B: Methods of DNA extraction, primer design and phylogenetic analyses To extract DNA, each thawed filter was unfolded into a 50 ml Falcon tube to which 2 ml of lysis buffer was added, followed by 100 µl of lysozyme (125 mg/ml) and 20 µl of RNase A (10 µg/ml). The tubes were sealed with parafilm and placed in an incubator at 37°C for 1hr. 100µl of Proteinase K (10 mg/ml) and 100µl of SDS (20%) were added before another incubation at 55°C for 2hr. We then removed protein using protein precipitation reagent (MPC) and centrifugation at 10 000G at 4°C for 10 min. DNA was precipitated in isopropanol, washed with 70% ethanol, and then re-suspended in 10 mM Tris buffer. To design our primers, we aligned all sequences of Scenedesmus ITS2 retrieved from the ITS2 Database (Schultz et al. 2006) using the Geneious software (Drummond et al. 2012). The forward (5’-catgtctgcctcagcgtcg-3’) and the reverse (5’-ggtagccttgcctgagctca-3’) primers were located in the conserved 5.8S and 28S rDNA regions, respectively. PCR reactions (25 μl total volume) contained 0.5 μM each primer, 1 X Phire Reaction Buffer containing 1.5 mM MgCl2 (Finnzymes Thermofischer Scientific), 0.2 mM dNTPs, and 0.5 U of Phire Hot Start II DNA Polymerase (Finnzymes Thermofischer Scientific). Cycling conditions involved an initial 3 min denaturing step, followed by 30 cycles of 5 seconds at 98 °C, 5 s at 49 °C, and 10 s at 72 °C, and a final elongation step of 1 min at 72 °C. Reverse primer were barcoded with a specific IonXpress sequence to identify samples. PCR products were purified using QIAquick Gel Extraction Kit (Qiagen), quantified using Quantifluor dsDNA System (Promega), pooled at equimolar concentration, and sequenced using an Ion Torrent PGM system on a 316 chip with the Ion Sequencing 200 kit as described in Sanschagrin and Yergeau (2014). 7 For the phylogenetic analysis of the ITS2 sequence data, raw sequences were processed using Mothur (Schloss et al. 2009): sequences shorter that 148 bp (expected size of the ITS2 amplicon) were removed. Low quality sequence data was trimmed from the 3’ end to a final length of 148 bp. To further simplify the phylogenetic analyses, we binned the sequences to different taxonomic groups. To do so, we clustered sequences using CD-HIT-EST algorithm on the CD-HIT server (Li and Godzik 2006) and a single representative sequence of each of the 298 identifed clusters was searched against the NCBI nucleotide database using nucleotide BLAST (Altschul et al. 1997) and assigned to a known genus based on the top BLAST hit. All sequences assigned to a genus were then separately aligned with MUSCLE v3.8.31 (Edgar 2004). In cases where there were >1500 sequences to align (the case for Chlamydomonas, Coelastrum, Desmodesmus, Oedogonium and Yamagishiella), we limited MUSCLE to 1 iteration. 8 Appendix C: Details of the phytoplankton morphospecies community responses in the mesocosm experiment based on microscopic identification With respect to the phytoplankton morphospecies (taxonomic) composition, Synedra sp. and Cyclotella glomerata were the dominant diatom morphospecies and both persisted the longest in all treatments. Chlorophyte replacement following perturbation resulted mainly from growth of Scenedesmus spp., and while they were present before acidification, they became dominant and more diversified following acidification. Of the ten Scenedesmus species, Scenedesmus ecornis, followed by Scenedesmus incrassatulus, were dominant. Other chlorophyte species that had been rare or undectable prior to perturbation became more important following it, including the two filamentous taxa of Mougeotia sp. and Ulothrix sp. It is important to note that the dominant chlorophyte, Scenedesmus spp. was identified using microscopic identification and taxonomic keys based on morphology (Prescott 1964, 1982; Whitford and Schumacher 1984). Subsequent molecular analyses classified them as Desmodesmus spp. There were two exceptions in which a Scenedesmus species was also detected by molecular methods, although they could not be identified to species because their ITS2 region had not previously been sequenced. Desmodesmus was considered to be a subgenus of Scenedesmus until the late 1990’s when studies sequencing ITS2 determined that they should be classified as a separate genus (Kessler et al. 1997; An et al. 1999). These two genera are morphologically cryptic and also highly plastic (Lürling 2003; Johnson et al. 2007), further complicating their identification through microscopy. The first axis of the PRC (Table C1, Fig. C1) was significant (P =0.005), indicating that while there was little community variation before perturbation, post-acidification a 9 differentiation of the Treatments from the Control occurred. The Pulse treatment diverged less from the Control than the Press, and a differentiation between the two perturbed treatments began about one week after acidification (Fig. C1). Some species responded negatively, including all cryptophytes, indicating that they declined in the treated mesocosms relative to the Controls. Most species that reacted positively showed a mild reaction (score <1), and most were chlorophytes. The PRC scores by morphospecies are given in Table C1, along with the full names for each abbreviation (Fig. C1). References: An, S., T. Friedl, and E. Hegewald. 1999. Phylogenetic Relationships of Scenedesmus and Scenedesmus-like Coccoid Green Algae as Inferred from ITS-2 rDNA Sequence Comparisons. Plant Biology 1: 418–428. Johnson, J., M. Fawley, and K. Fawley. 2007. The diversity of Scenedesmus and Desmodesmus (Chlorophyceae) in Itasca State Park, Minnesota, USA. Phycologia 46: 214–229. Kessler, E., M. Schafer, C. Hummer, A. Kloboucek, and V. Huss. 1997. Physiological, biochemical and molecular characters for the taxonomy of the subgenera of Scenedesmus (Chlorococcales, Chlorophyta). Botanica Acta 110: 244–250. Lürling, M. 2003. Phenotypic plasticity in the green algae Desmodesmus and Scenedesmus with special reference to the induction of defensive morphology. Annales de LimnologieInternational Journal of Limnology 39: 85–101. Prescott, G. 1964. How to Know the Freshwater Algae, WM.C Brown. Dubuque, Iowa. Prescott, G. 1982. Algae of the Western Great Lakes Area. Otto Koeltz Science Publishers, Koenigstein. Whitford, L., and G. Schumacher. 1984. A Manual of Fresh-water Algae, Sparks Press, Raleigh, NC. 10 Table C1: Abbreviated and full names of phytoplankton morphospecies identified microscopically across all of the experimental communities and their scores associated with the PRC (Fig. C1). Class Chlorophytes Abbreviation Ank.con Chl.sp Clo.sp Coe.sp Cos.sub Cos.ten Eud.sp Glo.sp Mou.sp Ooc.lac Ooc.sol Ped.tet Sce. arm Sce.inc Sce.qua Sch.sp Sph.sch Ulo.sp Wes.bot Full name Ankistrodesmus convulatus Chlamydomonas sp. Closterium sp. Coelastrum sp. Cosmarium subalatum Cosmarium tenue Eudorina sp. Gloeocystis sp. Mougeotia sp. Oocystis lacustris Oocystis solitaria Pediastrum tetras Scenedesmus armatus Scenedesmus incrassatulus Scenedesmus quadricauda Schroderia sp. Sphaerocystis schroeteri Ulothrix sp. Westella botryoides Score -0.0428 -1.9820 -1.5160 3.5030 0.9166 -0.1801 0.0736 -3.1230 -0.7456 -1.3840 0.0363 -0.4765 1.9640 -0.4970 0.0336 1.4640 -0.5967 -1.5600 -1.2790 Diatoms Tab.sp Tabellaria floculosa -3.2850 Cryptophytes Cry.bor Cry.ero Rho.sp Cryptomonas borealis Cryptomonas erosa Rhodomonas sp. 2.2650 0.9677 1.5250 Chrysophytes Chr.sp Chromulina sp. -1.1390 Cyanobacteria Osc.ute Oscillatoria utermoehlii 0.5863 11 Figure C1: Axis 1 of the PRC analysis for the morphospecies community responses to the treatments. The grey horizontal line indicates the Control treatment; the dashed line indicates the Pulse treatment and the black line the Press treatment. Larger treatment effects are indicated by a more distant line from the Control. Response strengths of responsive morphospecies are shown along the right side according to the species abbreviations and values in Table C1. Only morphospecies with a species score greater than +/- 0.5 in a PRC are considered responsive (Van den Brink and Ter Braak 1999) and are shown. Because the response curves of our two treatments are shown as negative, all the species displayed on the negative side of the y-axis have a positive response to the treatment and vice versa. 12 Appendix D: Details of the phylotypes identified in the experiment Table D1 : Phytoplankton genera and number of associated phylotypes identified using ITS2 sequencing. Genus Chlamydocapsa Chlamydomonas Coelastrum Desmodesmus Gonium Micractinium Oedogonium Pandorina Scenedesmus Sorastrum Yamagishiella Total Number of phylotypes 1 16 6 14 2 3 16 2 5 2 6 73 13 Figure D1: Phylogenetic analyses of algal ITS2 phylotypes associated to the genera : (A) Coelastrum and (B) Oedogonium. Phylotypes observed in the present study are indicated bold lettering. Boxes next to phylotype names indicate for which phylotypes either negative (black) or positive (gray) responses to the Pulse (left sub-box) and the Press (right sub-box) were observed. Bootstrap values >60 are represented by black circles on branches. (A) 14 (B) 15 Appendix E: Responses of the genotypes (97% identity clustering) Figure E1: Histograms showing the relative abundances of observed genotypes within (A) Dcun-036 and (B) Csp-070 through time across treatments (columns) for each replicate mesocosms (rows). Note, only two Control replicates could be analysed genetically (see Methods). Numbers above bars are absolute number of sequences for the entire phylotype. (A) (B)