jbi12615-sup-0001-AppendixS1
advertisement

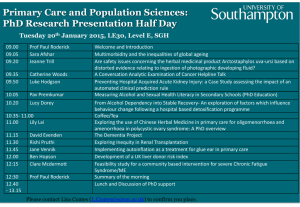
Bradbury, Tapper, Coates, Hankinson, McArthur and Byrne; “How does the post-fire facultative seeding strategy impact genetic variation and phylogeographic history? The case of Bossiaea ornata (Fabaceae) in a fire-prone, mediterranean-climate ecosystem”; Supplementary Files How does the post-fire facultative seeding strategy impact genetic variation and phylogeographic history? The case of Bossiaea ornata (Fabaceae) in a fire-prone, mediterranean-climate ecosystem. Donna Bradbury, Sarah-Louise Tapper, David Coates, Maggie Hankinson, Shelley McArthur and Margaret Byrne * Journal of Biogeography Appendix S1. Methods and results of nuclear population structure analysis using STRUCTURE 2.3 (Pritchard et al., 2000; Falush et al., 2003) and TESS 2.3.1 (Chen et al., 2007) in Bossiaea ornata using eight nuclear microsatellite markers. STRUCTURE: Optimal K was determined based on the ΔK method of Evanno et al. (2005), implemented in STRUCTURE HARVESTER 0.6.94 (Earl & vonHoldt, 2012), and by assessing the slope of lnP(K) with increasing K value. TESS: The most likely K was determined by obtaining the deviance information criterion (DIC) scores for all 100 runs, and plotting the mean of the 10% of runs with the lowest DIC scores against Kmax. Optimal K was determined when the graph reached a plateau. The Greedy algorithm within CLUMPP (Jakobsson & Rosenberg, 2007) was used to determine congruence of results among runs (H’) and to summarise the optimal alignment, and DISTRUCT (Rosenberg, 2004) was used to plot a bar graph of individual and population membership q-values. Bradbury, Tapper, Coates, Hankinson, McArthur and Byrne; “How does the post-fire facultative seeding strategy impact genetic variation and phylogeographic history? The case of Bossiaea ornata (Fabaceae) in a fire-prone, mediterranean-climate ecosystem”; Supplementary Files Bradbury, Tapper, Coates, Hankinson, McArthur and Byrne; “How does the post-fire facultative seeding strategy impact genetic variation and phylogeographic history? The case of Bossiaea ornata (Fabaceae) in a fire-prone, mediterranean-climate ecosystem”; Supplementary Files Appendix S2. Genetic diversity characteristics of eight nuclear microsatellite loci used to genotype 290 individuals of Bossiaea ornata. Locus Bo6G8 BoAg3E2 BoAg1E12 Bo5A10 Bo1D6 BoAg3E4 Bo5D1 Bo3D2 All loci a AT 26 35 28 33 26 30 25 38 Size 150-210 246-312 116-222 215-269 205-243 266-326 185-239 226-275 HO 0.632 (0.044) 0.489 (0.036) 0.691 (0.034) 0.867 (0.020) 0.773 (0.031) 0.845 (0.026) 0.797 (0.032) 0.807 (0.027) HE 0.795 (0.019) 0.804 (0.018) 0.737 (0.029) 0.842 (0.014) 0.734 (0.030) 0.873 (0.008) 0.831 (0.018) 0.853 (0.009) F 0.209 (0.049)a 0.384 (0.051)a 0.056 (0.036)a -0.032 (0.023) -0.058 (0.020) 0.033 (0.027) 0.038 (0.038)a 0.055 (0.027)a Significant heterozygote deficit (p<0.05) Bootstrapped 95% confidence interval in parentheses AT – total number of alleles Size Range – allele size range (bp) HO – observed heterozygosity HE – expected heterozygosity F – fixation index S – private alleles FST –locus specific population differentiation, corrected for the presence of null alleles by the ENA method Null – mean null allele frequency, range in square parentheses. Standard errors are in round parentheses unless otherwise indicated. b S 7 10 9 9 7 2 4 11 FST 0.044 0.067 0.100 0.059 0.105 0.039 0.040 0.043 0.062 (0.049-0.081)b Null 0.09 [0.00-0.23] 0.18 [0.04-0.31] 0.04 [0.00-0.14] 0.00 [0.00-0.04] 0.00 [0.00-0.08] 0.02 [0.00-0.13] 0.03 [0.00-0.14] 0.03 [0.00-0.12] Bradbury, Tapper, Coates, Hankinson, McArthur and Byrne; “How does the post-fire facultative seeding strategy impact genetic variation and phylogeographic history? The case of Bossiaea ornata (Fabaceae) in a fire-prone, mediterranean-climate ecosystem”; Supplementary Files Appendix S3. Pairwise FST values among 15 natural populations of Bossiaea ornata, based on eight nuclear microsatellite markers. Significant values are highlighted in bold (p<0.05 after Bonferroni correction). SADD NORB SADD COLN COLS BEAU KIRP GREE NANN BAUD MARG MANN SCRE MANE PEER COLN 0.018 COLS BEAU KIRP GREE NANN BAUD MARG MANN SCRE MANE PEER MUIR 0.035 0.032 0.046 0.026 0.017 0.024 0.097 0.087 0.062 0.082 0.032 0.065 0.085 0.032 0.044 0.060 0.042 0.037 0.034 0.099 0.090 0.073 0.092 0.045 0.085 0.097 0.046 0.055 0.023 0.036 0.049 0.121 0.103 0.083 0.100 0.053 0.089 0.107 0.038 0.027 0.016 0.043 0.108 0.108 0.074 0.103 0.036 0.080 0.108 0.045 0.026 0.057 0.122 0.114 0.105 0.117 0.060 0.104 0.123 0.017 0.023 0.103 0.094 0.067 0.083 0.037 0.068 0.096 0.029 0.082 0.070 0.056 0.068 0.023 0.056 0.078 0.073 0.064 0.045 0.063 0.027 0.042 0.068 0.015 0.065 0.046 0.058 0.060 0.047 0.070 0.020 0.062 0.041 0.065 0.086 0.020 0.057 0.036 0.065 0.012 0.085 0.042 0.040 0.072 Bradbury, Tapper, Coates, Hankinson, McArthur and Byrne; “How does the post-fire facultative seeding strategy impact genetic variation and phylogeographic history? The case of Bossiaea ornata (Fabaceae) in a fire-prone, mediterranean-climate ecosystem”; Supplementary Files References Chen C., Durand E., Forbes F. & François O. (2007) Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Molecular Ecology Notes, 7, 747–756. Earl D.A. & vonHoldt B.M. (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4, 359–361. Evanno G., Regnaut S. & Goudet J. (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14, 2611–2620. Falush D., Stephens M. & Pritchard J.K. (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics, 164, 1567–1587. Jakobsson M. & Rosenberg N.A. (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23, 1801–1806. Pritchard J.K., Stephens M. & Donnelly P. (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. Rosenberg N.A. (2004) DISTRUCT : a program for the graphical display of population structure. Molecular Ecology Notes, 4, 137–138.