There are several ways to use the Heat Kernel Signature to compute
advertisement

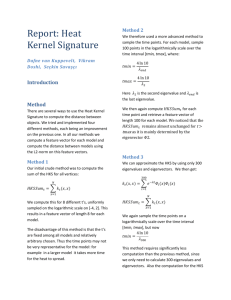
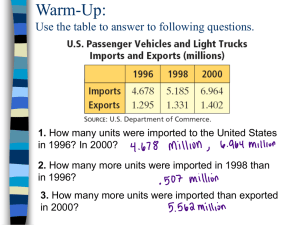
Report: Heat Kernel Signature Dafne van Kuppevelt, Vikram Doshi, Seçkin Savaşçı Introduction Method There are several ways to use the Heat Kernel Signature to compute the distance between objects. We tried and implemented four different methods, each being an improvement on the previous one. In all our methods we compute a feature vector for each model and compute the distance between models using the L2-norm on this feature vectors. Method 1 Our initial crude method was to compute the sum of the HKS for all vertices: 𝑁 𝐻𝐾𝑆𝑆𝑢𝑚𝑡 = ∑ 𝑘𝑡 (𝑥, 𝑥) 𝑥=1 We compute this for 8 different t’s, uniformly sampled on the logarithmic scale on [-4, 2]. This results in a feature vector of length 8 for each model. The disadvantage of this method is that the t’s are fixed among all models and relatively arbitrary chosen. Thus the time points may not be very representative for the model: for example in a larger model it takes more time for the heat to spread. Method 2 We therefore used a more advanced method to sample the time points. For each model, sample 100 points in the logarithmically scale over the time interval [tmin, tmax], where: 𝑡𝑚𝑖𝑛 = 4 ln 10 𝜆𝑒𝑛𝑑 𝑡𝑚𝑎𝑥 = 4 ln 10 𝜆2 Here 𝜆2 is the second eigenvalue and 𝜆𝑒𝑛𝑑 is the last eigenvalue. We then again compute 𝐻𝐾𝑆𝑆𝑢𝑚𝑡 for each time point and retrieve a feature vector of length 100 for each model. We noticed that the 𝐻𝐾𝑆𝑆𝑢𝑚𝑡 remains almost unchanged for t > tmax as it is mainly determined by the eigenvector 𝛷2. Method 3 We can approximate the HKS by using only 300 eigenvalues and eigenvectors. We then get: 300 𝑘𝑡 (𝑥, 𝑥) = ∑ 𝑒 −𝜆𝑡 𝛷𝑖 (𝑥)𝛷𝑖 (𝑥) 𝑖=1 𝑁 𝐻𝐾𝑆𝑆𝑢𝑚𝑡 = ∑ 𝑘𝑡 (𝑥, 𝑥) 𝑥=1 We again sample the time points on a logarithmically scale over the time interval [tmin, tmax], but now 4 ln 10 𝑡𝑚𝑖𝑛 = 𝜆300 This method requires significantly less computation than the previous method, since we only need to calculate 300 eigenvalues and eigenvectors. Also the computation for the HKS of one vertex takes less time, namely O(300) instead of O(N). Method 4 In the previous methods, we sum over all vertices to get the signature of the complete model. This means that points on the model are not compared to corresponding points on another model. To accomplish this in the fourth method we find 200 distinct feature points on the model. These feature points have the highest local maxima, using multi scaling: 𝑡=𝑚𝑎𝑥 𝑑[𝑡𝑚𝑖𝑛,𝑡𝑚𝑎𝑥] (𝑥, 𝑥 ′) 𝑘𝑡 (𝑥, 𝑥) − 𝑁 ∑ 𝑘𝑡 (𝑥, 𝑥 ) 𝑡=𝑡𝑚𝑖𝑛 𝑖=1 = ∑ − 𝑘𝑡 (𝑥′, 𝑥′) ∑𝑁 𝑖=1 𝑘𝑡 (𝑥′, 𝑥′) This results in a feature vector of length 200 for each model. Implementation As soon as we became confident and we had a consensus about the ideas in our reference paper, we started the implementation. Rather than implementing all sequence together, we divided our implementation into 4 stages: the Initial Parser is where we compute Laplacian matrices from 3D models and the eigenvalues and -vectors from those Laplacian matrices; using these eigenvalues and vectors, we calculate the HKS feature vectors in the HKS Calculation stage; we focused on matching and distance calculations and the resulting performance measures in Retrieval ; in the Visualization stage we visualize our results of the Heat Kernel Signature calculations. Initial Parser The Initial parser contains the operations that take much time, but luckily only need to be done once. These include the computation of the Laplacian matrices and the corresponding eigenvalues and eigenvectors. The main challenge in this stage is to find efficient techniques to perform this calculations and make sure that we can use the results of this stage in later stages easily. a. Computing Laplacian Matrices We tried to write the code for the computations ourselves as much as possible, without using external libraries. Our goal was to implement a method that parses 3D models and outputs the Laplacian matrices to text files, one by one. Our first successful implementation was using integers for keeping each element of the resulting Laplacian matrix; also it was giving the output in human readable format. But tests showed that it was slow and too much space consuming. Our test cases showed that if we wanted to parse every model, it would take more than 2 hours to finish. Also for the largest models (in terms of vertex count), it would go beyond x86 address space. To be more precise, our largest model has 45000 vertices, which results in a matrix data structure of size 45000 ∗ 45000 ∗ 4 𝐵 = 7.54 𝐺𝐵 which is greater than our accessible address space (2 GB, OS restriction) in Windows, also greater than 232 Bytes which we can address in x86. We came up with a solution, namely reducing our matrix and using a bit to represent each element of the matrix. The Laplacian matrix is a sparse symmetric matrix, where one can compute the diagonal values by traversing the corresponding row or column. We therefore decided to store only the lower triangular part of it and we also omitted the diagonal part because it can be computed by row traversal. The resulting data is a lower triangular matrix without the diagonal part. By definition of the Laplacian matrix, elements in this part can only take the value -1 or 0. We replaced -1s with 1s, so each element could be stored using a bit. If the program calls an upper triangle’s element we send the lower part correspondence, and if the diagonal part element is needed, we compute it on the fly. By using this schema, we were able to parse all files in 2 minutes including the largest ones that we couldn’t parse in the previous method. To be more precise again, we then needed only 45000∗44999 2 𝑏𝑖𝑡𝑠 = 120 𝑀𝐵 space for each file. Result was a ~64X compression. Figure 1: Simple Model; Table 1: Our First Implementation; Table 2: Compressed Implementation The next decision we had to make was what the storage format we would use for these Laplacian matrices. Our first implementation was giving a human readable text file, in which we converted the values to strings and added white spaces and line feeds for readability. However, such formatting increases the parsing time. In addition, we had to implement a loader method that loads these matrices into memory when we need it. We eventually decided to serialize our matrix data to files. With this decision, our resulting files were no longer human-readable, but we could simply deserialize it in our program to use it for further calculations. b. Computing Eigen Vectors & Values Computing eigen values and vectors is a challenging problem, because of its high complexity. As we stated before, at first we tried to implement our own eigen decomposer. But after some considerable effort, we didn’t manage to implement a working version; so we decided to use a 3rd party library for eigen decomposition. In literature, there are specialized techniques - such as Lanczos algorithm- for different kind of matrices, so we had to find a good library that fits to our input laplacian matrices which are sparse symmetric matrices. Search for a library that had a specialized eigen decomposer for our type of matrices had failed, so we started to try every library that we could find on the web, and evaluated them. library Spec License Time Memory Math.NET C# x86 Sequential LAPACK xGPL 20+ mins Average DotNumerics C# x86 Sequential LAPACK port xGPL 2 mins Average CenterSpace C# x86 Sequential LAPACK $1295 30+ mins Good Eigen C++ x86 Sequential UNIQUE xGPL 2 hours Good Table 3: Evaluation results for 2000 vertex model After the evaluation of some libraries, we decided use DotNumerics library. Using DotNumerics, we found a solution to eigen decomposition, but it had some drawbacks in our project: We must ignore the largest ~40 matrices: DotNumerics uses double precision for storing matrix elements, although DotNumerics have a symmetric matrix data storage class, it results in huge memory need which is more than x86 architecture can provide for the largest matrices. We need to create a library copy of our laplacian matrices during parsing, because DotNumerics uses LAPACK routines under the hood and that means we cannot use our bitwise matrix format as an input for Eigen Decomposition Methods. We also continued using serialization for outputs of the library. Computed eigen values and vectors are serialized to corresponding files. c. An experimental Approach for Future Implementations Although we used DotNumerics thru our project and ignore the largest matrices, we came up with a prototype for future work. Our prototype is written in C++ rather than C#, uses Intel Math Kernel Library©, Boost and a tweaked version of Armadillo libraries. Prototype parser runs on x64 architecture, and it takes only 10 seconds to compute eigen values and eigen vectors of a 2000 vertex model( To remind you, DotNumerics does the same job in 2 mins). We had two main reasons for developing this prototype. At first, today’s CPUs are mostly x64 capable, and we wanted to see how better it could be if we run the same implementation on x64. Secondly, an x86 Windows OS limits the available memory to 2 GB per process; but we need more memory than that if we want to parse the largest matrices. However we must warn the reader about performance. We tested this prototype on a machine that has 3GB main memory, and rest of the data is stored on virtual memory. In largest matrices disk I/O and, as a result of it, the computation time increased dramatically. HKS Calculation Retrival Visualization & Proof of correctness Visual proof of our implementation’s correctness was a must, and we provided both a 3d model visualizer implementation and graphs of our retrieval results. For the 3d model visualizer, we used XNA framework. Our implementation reads 3d model and corresponding HKS value file and draws a 3D scene. One can navigate in this scene using an XBOX controller. Also if provided, you can see the different results of model in different time values. Performance Conclusion