ADB-ADB2-Atkins20111143-RR
advertisement

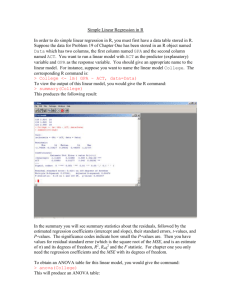
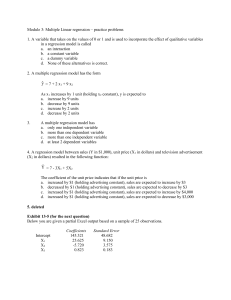
Longitudinal Count Regression 1 Technical Appendix This technical appendix covers several issues that are important in fitting GLMMs but also require more statistical background than is assumed in the main body of the text. Brief Overview of Optimization in GLMMs A critical issue for more complex statistical models is estimation. Considering the models described in equations 1-3 (and GLMMs generally), what method can be used to estimate the various parameters (i.e., fixed and random-effects)? Linear regression is often referred to as ordinary least squares regression, which describes the estimation strategy used for linear regression. Least squares estimation is directly parallel to solving simple algebra problems: There is an equation with one or more unknowns that can be solved directly. However, as soon as we move beyond OLS regression, many statistical models no longer have a “closed form” solution to estimation, and iterative fitting algorithms are required. GLMMs can be estimated via maximum likelihood (ML) estimation or Bayesian Markov chain Monte Carlo (MCMC) estimation, which are both considered here briefly. Software implementations of ML for linear mixed models are now fairly quick and accurate except for special circumstances (e.g., cross-classified models or extremely large datasets). However, with GLMMs there is an added layer of complexity that comes with the non-normal outcome. Specifically, to solve the likelihood equation, it is necessary to integrate over the random-effects, which is far more challenging with a non-identity link function that connects the left and right-hand side of the model. Currently, various estimation methods are used for parameters in GLMMs. Here we provide a general description aimed at applied issues, but further details can be found in Raudenbush and Bryk (2002) and Hedeker and Gibbons (2006). An excellent, article length overview of GLMMs that touches on estimation issues and Longitudinal Count Regression 2 software is Bolker et al. (2009), though examples are related to ecology. The earliest methods were called marginal quasilikelihood (MQL) and penalized quasilikelihood (PQL), but both MQL and PQL have been shown to lead to biased estimates, especially for variance terms (see, e.g., Rodriguez & Goldman, 2001). Moreover, the approximations used by these methods did not allow the use of deviance tests to compare models. PQL is still an option in some software packages (see Table 1 in Bolker et al., 2009). Currently, most software packages for GLMM use either a Laplace estimation procedure or adaptive Gaussian quadrature (AGQ). Both are more accurate than either MQL or PQL and yield accurate likelihood statistics that can be used for model comparison purposes. When available, the AGQ procedure is more accurate than estimation using Laplace estimation; however, AGQ can be quite slow, particularly with more than one random-effect (see, e.g., Rabe-Hesketh & Skrondal, 2008; Raudenbush & Bryk, 2002). In addition, to use AGQ it is necessary to specify a number of quadrature points, and greater accuracy can be achieved with more quadrature points. But, again, model fitting time increases as quadrature points increase. Thus, there is a clear trade-off between speed of estimation and accuracy. Particular for smaller datasets or complex models, estimation methods should be scrutinized. An alternative approach to estimation is to use Bayesian Markov chain Monte Carlo (MCMC) methods, which are growing in popularity as computing power has reduced many of the historically challenging issues in fitting such models. Bayesian models do have several fundamental differences from the more common statistical methods (often referred to as frequentist methods). Bayesian models explicitly incorporate prior information into the model, which has led to criticisms of Bayesian methods as subjective. It is possible to include specific information in prior distributions (e.g., the effect of a given parameter should lie in a specific Longitudinal Count Regression 3 interval) often called “informative priors.” However, in applied research it is far more common to use uninformative priors. These imply no practical preference for any specific value of a parameter over another. Moreover, except in small datasets, uninformative priors contribute a relatively small amount to the resulting estimates as compared to the data itself (Gelman & Hill, 2007). A second difference is that Bayesian methods typically use MCMC estimation. MCMC is a simulation-based estimation procedure, which has shown to be very accurate under a wide array of conditions (for an overview of Bayesian methods for GLMM, see Draper, 2008, or for a general introduction to Bayesian methods for social science, see Lynch, 2007). Although ML fitting procedures are iterative, there is always a convergence criterion at which point the algorithm stops. With MCMC estimation there is no convergence criterion, and the data analyst specifies a number of iterations, at which point the analyst must ascertain whether the simulations have converged to appropriate, final estimates. Although this aspect of Bayesian methods has also been somewhat controversial, there is a general consensus on both tools and guidelines for judging when results have converged (see, discussion of convergence issues in Draper, 2008, and Gelman & Hill, 2007). Finally, MCMC estimation typically leads to a sample of estimates for each parameter, resulting from the simulation (e.g., 1,000 draws from the simulation for each parameter might be saved for analysis). Somewhat similar to bootstrapping, these simulated draws from the posterior distributions of parameters can be summarized by their mean and confidence intervals. Although this quick overview of Bayesian methods may sound quite different from what occurs with more common statistical procedures (and ML for GLMMs), results for simpler models are often highly similar if not identical across Bayesian and frequentist approaches (Gelman & Hill, 2007). Thus, somewhat similar to AGQ, MCMC Longitudinal Count Regression 4 estimation is highly accurate though more time consuming than alternatives; however, one notable difference with ML estimation generally is that MCMC estimation has been shown to be accurate (i.e., point estimates and appropriate coverage of CI) even in small sample sizes (see, e.g., discussion in chapter 13 of Raudenbush & Bryk, 2002). Conditional vs. Marginal Fixed-Effects: Random-Effects and Link Functions in GLMM With a linear mixed model (LMM) the link function is the identity function, which practically means there is no link function (i.e., it is similar to multiplying something by one, nothing changes). In the main body of the text, we considered the implications of this for interpreting the fixed-effects, but it also has important implications for the random-effects, which in turn affects predictions from GLMM and directly relates to the distinction between marginal coefficients (sometimes called population-average models) vs. conditional coefficients (or unitspecific models). Most of the material in this section of the appendix is taken from Raudenbush and Bryk (2002; chapter 10), Raudenbush (2008), Breslow and Clayton (1993), and Heagerty and Zeger (2000). Let’s return to the full model from our initial Poisson GLMM: log(E[RAPIti]) = b0 + b1Male + b2Time + b3Male*Time + u0i + u1iTime (A1a) where subscripts are the same as earlier. All error terms are assumed normally distributed with a mean of zero and unknown variance. If equation A1a represented an LMM, then the randomeffects (i.e., subject specific effects) have mean zero on the scale of the linear predictor (i.e., right-hand side of equation) as well as the scale of the outcome (i.e., left-hand side of equation). Because of this, they do not contribute anything to the average predictions from the model. For example, simple slopes for interpreting the interaction of Male and Time do not need to include Longitudinal Count Regression 5 the random-effects, as the mean of zero and identity link function means that our predictions are averaging over the random-effects distributions. However, with GLMMs that have a non-identity link function, this relationship changes. The right hand side of the equation is considered the linear predictor in the language of GLMM, and it is connected to the outcome via a link function. For our count regression models, this is the log link, and we can make this explicit via: E[RAPIti] = exp(b0 + b1Male + b2Time + b3Male*Time + u0i + u1iTime) (A1b) With equation A1b the error terms still have a mean of zero, but only on the linear predictor scale. The error terms do not have a mean of zero on the original scale of the RAPI because we have to exponentiate the random effects to get them back to the original scale of the RAPI. For example, this can be seen in aFigure 1, which plots the subject-specific effects for the intercept from the model above (i.e., random-effects centered around the fixed-effect intercept), on both the scale of the outcome (y-axis) and the scale of the linear predictor (x-axis). Note that the distributions are centered around the fixed-effect intercept. The histogram on top of the graph shows the (approximately) normally distributed intercept variance on the linear predictor scale, whereas the marginal histogram to the right shows the same distribution on the scale of the outcome (i.e., after the values have been raised to the base e). The solid black line shows the mean of the subject-specific effects on the linear predictor scale, which is simply the fixed-effect estimate (i.e., mean of the distribution on the top of the graph; 4.59 on the outcome scale). The dotted line shows the mean of the exponentiated subject-specific effects (i.e., mean of the distribution on the right of the graph; 6.63 on the outcome scale). Several points are worth noting here: (a) the skewed distribution on the outcome scale makes sense with what we know of the data, but (b) exponentiating the random effects means Longitudinal Count Regression 6 that there is no longer a mean of zero. In fact, this is the reason why the rate ratio of the fixedeffect intercept (RR = 4.2) notably underestimates the average value of the outcome for women at time equal to zero (M = 6.3). Breslow and Clayton (1993) and others have noted that predictions from a Poisson GLMM need to include both fixed and random-effects: E[Y ] = exp(XB + diag(ZDZ ') / 2) A2 which uses matrix notation to designate: X as a fixed-effect design matrix, B as a vector of fixedeffect coefficients, Z as a random-effect design matrix (and Z' as its matrix transpose), and D as a variance-covariance matrix of random-effects. Finally, exp is the exponentiate function (i.e., raise to the base e, the inverse link function for Poisson) and diag specifies the diagonal elements of the resulting matrix. The equation above was used to estimate the marginal predictions from the count submodel of the TLFB example, shown in Figure 7. For that example, the components of the equation took the following values: é ê ê ê ê X =ê ê ê ê ê ë 1 1 1 1 1 1 1 1 0 1 0 1 0 1 0 1 0 0 1 1 0 0 1 1 0 0 0 0 1 1 1 1 0 0 0 0 1 0 0 1 0 0 0 0 0 1 0 1 0 0 0 0 0 0 1 1 ù é 0.98 ù ú ê ú ú 0.08 ú ê ú ê 0.18 ú ú ú B = ê 0.46 ú ê ú ú 0.11 ú ê ú ê -0.10 ú ú ê 0.19 ú ú ë û û é ê ê ê ê Z =ê ê ê ê ê ë 1 1 1 1 1 1 1 1 0 1 0 1 0 1 0 1 ù ú ú ú é 0.25 -0.01 ù ú ú D =ê ú ë -0.01 0.04 û ú ú ú ú û Several of these matrices are straightforward: B is the estimated fixed-effects for the count submodel on the scale of the link function (i.e., not transformed to RR), and D is the estimated variance-covariance matrix of random-effects. For the TLFB example, the count submodel includes a random intercept and random slope for weekend, and thus, D is a two by two matrix. X is a design matrix for the fixed effects, in which the columns correspond to the: a) intercept, b) Longitudinal Count Regression 7 weekend indicator, c) gender indicator, d) fraternity / sorority indicator, e) weekend by gender indicator, f) weekend by fraternity / sorority indicator, and g) gender by fraternity / sorority indicator. The interaction columns (i.e., final three columns) are simply the result of multiplying the appropriate main effect columns (e.g., the final column is the result of multiplying columns c and d). The rows of matrix X correspond to the eight subgroups crossing weekend, gender, and fraternity / sorority. For example, the first row has only the intercept included and corresponds to the estimated mean for women on weekdays who are not in a sorority (i.e., the group taking zero values on all covariates). The final row represents men on weekends who are in a fraternity (i.e., the group taking values of one on all covariates). Because the model includes random effects for the intercept and weekend, Z contains those two columns of X corresponding to these effects. One final note is that for the over-dispersed Poisson model, the per-observation variance also must be included.1 Using these values of the matrices and the formula above, the estimated marginal predictions from the count submodel of the TLFB example are: é ê ê ê ê E[Y ] = ê ê ê ê ê ë 3.45 3.71 4.20 5.07 5.49 5.34 8.29 9.04 ù ú ú ú ú ú ú ú ú ú û 1 The per-observation random-effect in an over-dispersed Poisson is like the residual error term in OLS regression (or a linear mixed model). Equation A2 can be extended to include this term via: E[Y ] = exp(XB + (diag(ZDZ ')+ s 2 ) / 2) This was the version of the equation used for the TLFB, where sigma (i.e., the per-observation random effect) was equal to 0.26. Longitudinal Count Regression 8 Within rounding error, these are the values that are plotted in Figure 7. Beyond the consideration that predictions from GLMM must incorporate random-effects terms, this example also underscores that the interpretation of coefficients from GLMMs are somewhat different than their LMM counter-parts. In the statistical literature this distinction is often discussed as differences between population-average vs. unit-specific (e.g., Raudenbush & Bryk, 2002) or marginal vs. conditional (e.g., Heagerty & Zeger, 2000). As a concrete example, the model in equation 1 was re-fit using generalized estimating equations (GEE; Liang & Zeger, 1986). GEEs are an alternative class of statistical models that are also appropriate for longitudinal and clustered data. As opposed to GLMM, GEE models treat the correlations due to the clustering as a nuisance parameter, and GEE do not directly model subject-specific effects as in GLMM. For present purposes, their critical feature is that GEE coefficients have a marginal or population-average interpretation; GEE coefficients do average over the individual differences. Table A1 displays the raw coefficients (i.e., on log scale) from GEE and GLMM fits to the RAPI data. Between the two models, the coefficients are different to varying degrees. However, it can be readily shown that the GEE coefficients reflect approximate averages of the entire sample. For example, estimates of baseline drinking problems for women and men (e1.79 = 6.0 and e(1.79 + 0.28) = 7.9, respectively) are very close to the raw means in the sample. Because the GLMM coefficients are conditional on the random-effects distribution, they do not retain this average interpretation. Practically, what should we make of this? First, it is important to realize the distinction between conditional and marginal coefficients (and correspondingly between GLMM and GEE). It can be a bit startling to exponentiate the fixed-effect intercept of a Poisson GLMM and find that it is not that close to the mean in the raw data. Thus, for starters, it is important to realize Longitudinal Count Regression 9 there is a reason for this. Second, after realizing this, it can be tempting to think of marginal coefficients (and possibly GEE models) as being “correct” and conversely that GLMM are perhaps “incorrect.” Raudenbush (2008) casts the two models as having two different foci: GEE (or population-average coefficients) are more appropriate for questions about the sample as a whole, whereas GLMM (or unit-specific coefficients) are more appropriate for questions focused on individuals or distributions of individuals. However, it is likely that many times applied users would prefer to interpret output from GLMM as marginal coefficients, which is clearly not right. There are formulas to convert conditional coefficients from GLMM to marginal coefficients (e.g., see Heagerty & Zeger, 2000, which are automatically provided in the HLM software). For random intercept only models, these are straightforward, but for anything more complex, converting conditional to marginal is not trivial. For predictions from a GLMM (e.g., mean difference based on dichotomous covariate, or simple slopes), it is possible to use Equation A2 to effectively average over the random-effects distribution. The accompanying R code shows an alternative strategy of using a Monte Carlo simulation from the random-effects to yield marginal predictions. Given all this, should GEE be preferred (or, said another way, should GLMM be avoided) if the focus is on drawing conclusions about the sample as a whole? Unfortunately, it is not quite that straightforward. For simpler models, the decision between GEE and GLMM is a matter of convenience, and hence, GEE might be preferred for interpretational reasons. However, GEE have certain limitations: They make a stronger assumption about missing data and assume that time-varying covariates are not correlated over time. In addition, GLMMs estimate subject-specific effects, allowing the data analyst to examine the distribution of individual intercepts and slopes (e.g., what percentage of individuals are improving over time in Longitudinal Count Regression 10 a treatment study?), which at times may be substantively interesting. GEE treats the correlations due to clustering as a nuisance and does not estimate individual effects. Finally, there are broad discipline differences in the familiarity with the models generally. Mixed models have been much more prevalent in the social sciences, whereas GEE at this point is not as common. Extending linear mixed models to non-normal outcomes is likely building from an established foundation in the social sciences. In summary, our primary recommendation with respect to conditional versus marginal models is that GLMM users should familiarize themselves with the issues so that they might be informed users and correctly interpret their models. The citations noted throughout this section would be an excellent starting place. Longitudinal Count Regression 11 Table A1 RAPI Data Raw Coefficients for GEE and GLMM RRGEE RRGLMM Intercept 6.00 4.00 Male 1.32 1.22 Time 0.99 0.96 Male x Time 1.01 1.02 Longitudinal Count Regression 12 Figure Captions for Appendix Figure A1. Plot of subject-specific intercepts (i.e., random-effects centered around the fixedeffect intercept) on the scale of the linear predictor (x-axis) and outcome (y-axis). The solid line is the mean of the distribution on the linear predictor scale, whereas the dotted line is the mean on the outcome scale (i.e., after exponentiating all the values). Longitudinal Count Regression 13 Longitudinal Count Regression 14 Figure Captions for Extra Figures eFigure 1. Histograms and quantile-quantile plots of random effects from Poisson generalized linear mixed model fit to RAPI data, including over-dispersion term. eFigure 2. Plot of observed frequency counts along with predicted counts from Poisson GLMM (dotted line). Longitudinal Count Regression 15 Longitudinal Count Regression 16