tpj12203-sup-0012-Legends
advertisement

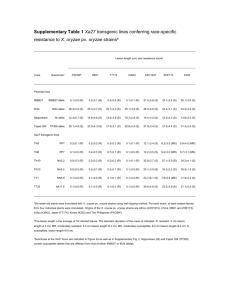
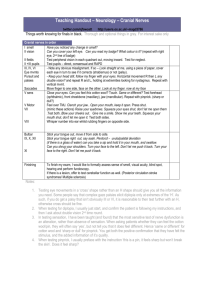
1 SUPPLEMENTAL FIGURE LEGENDS 2 Figure S1. Bc infection in plants grown at high density. A) Lesion diameters scored 3dpi on leaves 3 inoculated with a 5µl droplet of 5x105 Bc spore solution. I: lesion diameter < 1.5 mm, II lesion diameter ≥ 4 1.5 mm and ≤ 3 mm, III: lesion diameter > 3 mm (n > 20 leaves). B) Representative photographs of 5 inoculated leaves from plants grown individually (single) or at high-density canopies. 6 7 Figure S2. Leaf response to Bc-inoculation. Relative expression of PDF1.2 in Bc-inoculated petioles and 8 leaf laminas 2dpi (n = 3). Data represent means ± SE. Asterisk indicates significant difference (Student’s t- 9 test, p < 0.05). 10 11 Figure S3. Disease incidence of the constitutive defence overexpressors cpr1 and cev1 in control light 12 and low R:FR. A) Bacterial growth quantification 3dpi of Pst-inoculated leaves. Cfu = colony forming 13 units. B) Lesion diameters scored 3dpi on leaves inoculated with a 5µl droplet of Bc spore solution. As 14 average lesions size was much smaller in cev1 mutants (inset), different classes were used for Col-5 and 15 cev1 to analyse low R:FR effect. For Col-5 I: lesion diameter < 4 mm, II: lesion diameter ≥ 4 mm and ≤ 8 16 mm, III: lesion diameter > 8 mm. For cev1 I: lesion diameter < 1.5 mm, II: lesion diameter ≥ 1.5 mm and ≤ 17 3 mm, III: lesion diameter > 3 mm (n > 20 leaves). 18 19 Figure S4. Overlap in differentially expressed genes induced by 2h combined treatment of SA and low 20 R:FR and combined treatment of MeJA and low R:FR. The one overlapping gene is PHYTOCHROME 21 RAPIDLY REGULATED1 (PAR1), which just falls short on the statistical cut off used for this dataset (low 1 22 R:FR-induced adj.p = 0.005) and is a well-known low R:FR-induced gene (Roig-Villanova et al., 2007; 23 Sessa et al., 2005). 24 25 Figure S5. Heatmap of log2 Fold Change expression values of JAZ genes in MeJA treatment and MeJA 26 combined with low R:FR. All presented JAZ genes were significantly differentially expressed in both 27 treatments as compared to control except for JAZ1, which was not differentially induced by MeJA alone. 28 29 Figure S6. Volcano plots of JA and SA microarrays, representing the significance (log odds; the probability 30 that a gene is differentially expressed) and fold change for all genes. 31 32 SUPPLEMENTAL TABLE LEGENDS 33 Table S1. GO analysis of differentially expressed genes from SA- and MeJA microarrays. Adjusted p- 34 values for enriched GO categories are depicted for separately analysed subgroups from the venn 35 diagrams in Fig. 4). 36 Table S2. Gene IDs corresponding to Fig. 5C and D. 37 Table S3. SA-induced kinases repressed by low R:FR. 38 Table S4: Primer sequences (5’→3’) used for Real-Time RT-PCR. F: forward, R: reverse. 39 Table S5: Primer sequences (5’→3’) used for semi-quantitative RT-PCR. F: forward, R: reverse. 40 41 2 42 43 44 3