Properties_of_a_Ratio_EBV
advertisement

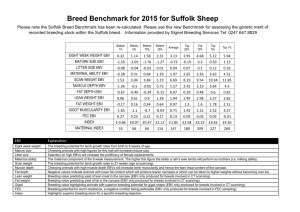
Properties of a Ratio EBV for Fat% Pete Sullivan and Gerrit Kistemaker Slopes of prediction, from genomic validation tests applied to percentages of Fat (F%) and Protein (P%) in milk are much higher than slopes of prediction for the corresponding yield traits; F, P and milk (M). These slopes of prediction, of current EBV from 4-years-ago GPA, are very close to the expected value of 1.00 for the yield traits but are much closer to 1.25 for F% and P%. The higher slope for F%, for example, indicates that GPA will under-predict genetic differences among young bulls, and that top young genomic bulls for F% will achieve much higher EBV after progeny testing than was predicted by their GPA prior to progeny testing. We have observed, and recently reduced, problems of low slopes of prediction for GPA that are linear functions of multiple traits (e.g. daughter fertility, calving ability, and indirect herd life), but since F% and P% are ratios they are thus non-linear functions, and the methods that improved GPA slopes of prediction for linear functions have not improved slopes of prediction for F% or P% (recently tested). Further, we see no difference in pattern regardless of which genetic prediction is used for the young bull (PA, DGV or GPA). Slopes of prediction for F% and P% are always much higher than for the corresponding yield traits (USA observes very similar pattern as well). To better understand why genetic differences for percentage (ratio) traits are always under-predicted, we generated a set of bias-corrected 4-years-ago PA for M and F yields. This approach was to eliminate both a) genomic evaluation and b) bias in yield trait evaluations as the explanations for bias in F%. The bias-corrected PA for M and F have slopes of prediction exactly equal to 1.00. F% was derived from these PA for F and M yield as follows: f% (f F) F , (m M) M where F =357 and M =9561 (Holstein phenotypic averages for base cows in 2010) and m, f and f% are EBV (PA) for M and F yield and F% respectively. The table below shows, in black text, correlations among the EBV in 2014, the bias-corrected PA in 2010 and the prediction residuals (e) between these two variables (i.e. EBV-PA) for each of M, F, and F%. Slopes of prediction (b(EBV,PA)) are also shown below the table. Correcting the slope for M and F did not correct the slope of prediction for F%, which was still very much higher (1.25) than 1.00, as we see in all of our applied validation results. We partitioned the respective contributions (C) of f and m EBV to the f% EBV as follows: f F F f % C(f ) C(m) (m M) (m M) M These contributions were derived for EBV, PA and the difference (e=EBV-PA). Correlations and slopes of prediction for the partitioned contributions and the product of the two are shown in red text in the Table. Slopes of prediction for C(f) and C(m) were close to 1.00, but for the product of the contributions ((f)*(m)) the slope was much lower at only 0.77. Different correlations between M and F for PA (0.63) versus EBV (0.39) and the C(f)*C(m) interaction effect provide some clues about the higher slope of prediction for F%. However, what can be done to generate unbiased f% from unbiased f and m evaluations (slope for f% closer to 1.00)? Are there general methods for ratio-defined traits that can help, modifications to the formula for f% to account for r(M,F), ...? Dairy Cattle Breeding and Genetics Committee meeting, Guelph, September 30, 2015 Page 1 of 2 EBV - PA PA EBV Correlations 2014 EBV MILK FAT FAT% C(f) 2010 PA C(m) (f)*(m) MILK FAT FAT% C(f) 2014 EBV - 2010 PA C(m) (f)*(m) MILK FAT FAT% C(f) C(m) MILK 1.00 FAT 0.39 1.00 FAT% -0.54 0.56 1.00 C(f) 0.34 1.00 0.60 1.00 C(m) -0.99 -0.39 0.54 -0.35 1.00 (f)*(m) -0.62 -0.60 0.03 -0.53 0.59 1.00 MILK 0.56 0.28 -0.25 0.26 -0.56 -0.32 1.00 FAT 0.32 0.55 0.22 0.55 -0.32 -0.34 0.63 1.00 FAT% -0.27 0.32 0.54 0.34 0.27 -0.02 -0.42 0.44 1.00 C(f) 0.30 0.56 0.24 0.56 -0.31 -0.33 0.59 1.00 0.48 1.00 C(m) -0.56 -0.28 0.24 -0.27 0.57 0.32 -1.00 -0.63 0.42 -0.60 1.00 (f)*(m) -0.43 -0.40 0.01 -0.38 0.42 0.41 -0.82 -0.82 -0.01 -0.79 0.80 1.00 MILK 0.83 0.28 -0.48 0.24 -0.82 -0.54 0.00 -0.04 -0.04 -0.04 0.00 0.04 1.00 FAT 0.26 0.83 0.53 0.83 -0.26 -0.50 -0.08 0.00 0.09 0.01 0.08 0.06 0.36 1.00 FAT% -0.50 0.50 0.90 0.54 0.49 0.05 -0.08 0.03 0.13 0.04 0.07 0.02 -0.55 0.58 1.00 C(f) 0.22 0.84 0.57 0.84 -0.22 -0.42 -0.07 0.01 0.10 0.02 0.07 0.05 0.31 0.99 0.62 1.00 C(m) -0.83 -0.29 0.49 -0.25 0.83 0.51 -0.01 0.03 0.05 0.03 0.01 -0.03 -0.99 -0.36 0.55 -0.31 1.00 (f)*(m) -0.43 -0.42 0.03 -0.35 0.40 0.85 0.12 0.10 -0.02 0.10 -0.12 -0.13 -0.61 -0.58 0.04 -0.49 0.57 1.00 1.00 1.25 1.03 1.02 0.77 b(EBV,PA) Dairy Cattle Breeding and Genetics Committee meeting, Guelph, September 30, 2015 (f)*(m) 1.00 Page 2 of 2