Consistency check on marker scores
advertisement

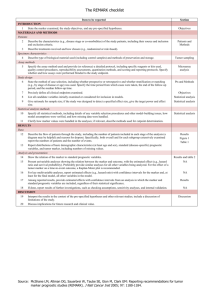
TITLE Bayesian QTL analyses using pedigreed families of an outcrossing species, with application to fruit firmness in apple AUTHORS MCAM Bink1, J Jansen1, M Madduri2, RE Voorrips2, C-E Durel3,4,5, AB Kouassi3,4,5 *, F Laurens3,4,5, F Mathis3**, C Gessler6, D Gobbin6***, F Rezzonico6,7, A Patocchi6,7, M Kellerhals7, A Boudichevskaia8****, F Dunemann8$, A Peil8, A Nowicka9, B Lata10, M Stankiewicz-Kosyl10, K Jeziorek11, E Pitera11, A Soska11, K Tomala11, KM Evans12,$*, F Fernández- Fernández 12, W Guerra13, M Korbin14, S Keller14, M Lewandowski14, W Plocharski14, K Rutkowski14, E Zurawicz14, F Costa15,16, S Sansavini15, S Tartarini15, M Komjanc16, D Mott16, A Antofie17$**, M Lateur17A Rondia17, L Gianfranceschi18, and WE van de Weg2 1 Biometris, Wageningen UR, Droevendaalsesteeg 1, P.O. Box 16, 6700 AA Wageningen, The Netherlands Plant Breeding, Wageningen UR, Droevendaalsesteeg 1, P.O. Box 16, 6700 AA Wageningen, The Netherlands 3 INRA, UMR1345 Institut de Recherche en Horticulture et Semences, SFR 4207 Quasav, Pres L’UNAM, F49071 Beaucouzé, France 4 Université d’Angers, UMR1345 Institut de Recherche en Horticulture et Semences, F-49045 Angers, France. 5 AgroCampus-Ouest, UMR1345 Institut de Recherche en Horticulture et Semences, F-49045 Angers, France. 6 Plant Pathology, Institute of Integrative Biology (IBZ), ETH Zurich, CH-8092 Zurich, Switzerland 7 Research Station Agroscope Changins-Wädenswil, ACW, Schloss 1, P.O. Box CH-8820, Wädenswil, Switzerland 8 Institute for Breeding Research on Horticultural and Fruit Crops, Julius Kühn-Institut, Pillnitzer Platz 3a, 01326 Dresden, Germany. 9 Department of Experimental Design and Bioinformatics, Warsaw University of Life Sciences – SGGW, 02-776 Warsaw, Poland 10 Laboratory of Basic Research in Horticulture, Faculty of Horticulture, Biotechnology, and Landscape Architecture, Warsaw University of Life Sciences SGGW, 02-776 Warsaw, Poland 11 Department of Pomology, Faculty of Horticulture, Biotechnology and Landscape Architecture, Warsaw University of Life Sciences – SGGW, 02-776 Warsaw, Poland 12 East Malling Research, New Road, East Malling, Kent ME19 6BJ, United Kingdom 13 Research Centre for Agriculture and Forestry Laimburg, Posta Ora – 39040 Vadena (BZ), Italy 14 Fruit Breeding Department, Research Institute of Horticulture, 96100 Skierniewice, Poland 15 Department of Fruit and Woody Plant Science, current Department of Agricultural Sciences, University of Bologna, Via Fanin 46, 40127 Bologna, Italy 16 Department of Genetics and Biology of Fruit Crops, IASMA Research and Innovation Centre, Foundation Edmund Mach, Via Mach 1, 38010 Trento, Italy 17 Walloon Agricultural Research Centre (CRA-W), Gembloux, Liroux 9, B-5030, Belgium 18 Department of Biosciences, University of Milan, Via Celoria 26, 20133 Milan, Italy 2 Current addresses: * Université Félix Houphhoët-Boigny, Unité de Formation et de Recherche (UFR) 'Biosciences', Laboratoire de Génétique, 22BP 582 Abidjan 22, Côte d’Ivoire ** Fabienne Mathis, VEGEPOLYS - Pôle de compétitivité , 7 rue Dixmeras - 49044 Angers cedex 01 France *** Tecan Group Ltd., CH-8708 Männedorf, Switzerland **** Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK), Corrensstr. 3, D-06466 Satdt Seeland Ot. Gatersleben $ Julius Kühn-Institut, Institute for Breeding Research on Horticultural and Fruit Crops, Erwin Baur Str. 27, D-06484 Quedlinburg, Germany $* Washington State University (WSU-TFREC), 1100 N. Western Avenue, Wenatchee, 98801- WA, USA $** Direction Générale Qualité et Sécurité - Métrologie Légale SPF Economie, P.M.E., Classes Moyennes et Energie, North Gate, Bd du Roi Albert II, 16, 1000 Bruxelles, Belgium CORRESPONDING AUTHOR Marco Bink Postal address Biometris, Wageningen University & Research centre PO Box 100, 6700 AC Wageningen The Netherlands Email marco.bink@wur.nl Electronic Supplementary Materials 3: Marker Map and Genotypes 3.1 Marker linkage map Figure ESM3.1: Integrated linkage map of 87 SSR loci on 17 chromosomes covering 1133 cM with map positions corresponding to those reported by Silfverberg-Dilworth et al. (2006). The map of Silfverberg-Dilworth et al. (2006) was over 1500 cM and 73% thereof was covered in this study. This figure was created with MapChart software (Voorrips 2002). 3.2 Marker genotypes The quality of the outcome of any QTL analysis is affected by the quality of the data. The FlexQTLTM software supports the use of high quality marker data by performing checks on the consistency of marker scores as well as by accounting for putative null-alleles. Moreover, FlexQTLTM assesses the polymorphism information content of the meiosis events for each marker locus. Consistency check on marker scores Consistency between marker and pedigree data is checked using a window of three successive generations. This is an iterative process of marker-by-marker checking of every individual. In case parental marker scores are lacking, consistency with grand-parental scores is still evaluated. In case of missing data, the FlexQTLTM software also performs imputation when the preceding and descending generations allow the deduction of an unambiguous marker genotype. Following this check, 41 (3%) progenies were removed being outcrossers and 15% of the initial marker scores was revised. Moreover, parentages were adjusted where appropriate (Evans et al. 2010). To date as many as 35 warnings are still unresolved. Null-alleles Loci that show just a single marker allele may be either homozygous or heterozygous with one so called null-allele. Null-alleles for co-dominant marker types, including SSR and SNP markers, occur regularly in the plant domain. Single marker alleles were coded as putative null-alleles to allow the FlexQTLTM software to impute either a (confirmed) null-allele or a homozygous allele conditional on an individual’s neighborhood set (parents, offspring and mates) (Fig ESM3.2) . The marker scores are made missing in case of ambiguous assignment. In several cases of ambiguous assignment of null-alleles (see below), allele dose was assessed through quantitative scoring of marker signals (Van Dijk et al. 2012), using other SSRmarkers from the same multiplex as reference. In our data set, 20% of the loci showed a single allele, 65% of which were imputed as homozygous, 16% as heterozygous null and 19% remained inconclusive. Moreover, 1200 (1%) locus/individual combinations did not show any marker because of having a null-allele in homozygous condition. RallsJan 213 null Delicious 207 213 Pinova 213 207 Fuji 207 null FuPi_025 FuPi_033 213 null 207 213 Figure ESM3.2: Imputation of a null allele for SSR locus CH03d01 based on an the marker scores of Fuji and its parents. Fuji shows a single marker allele that is present in only one of its parents, due to which Fuji cannot be homozygous. The other parent (Ralls Janet) shows just one marker allele too. Imputation of a null allele for Fuji and Rall Janet leads to a consistent inheritance pattern which holds over Fuji’s offspring (see FuPi_025). Polymorphism information content of marker meiosis events Part of the data of any locus will be of no use in monitoring recombination events due to the lack of marker scores, or due to individuals that are homozygous for this locus. For each locus, the FlexQTLTM software assesses the fraction of informative meiosis events, which ranged from 0.15 to 0.87 (Figure ESM3.3). Note that in diploid outbreeding species, the number of potentially informative meiosis is twice the number of individuals of the studypopulation that are no founders. Consistency among markers Consistency among markers is verified by comparing expected and observed numbers of double recombinants. For every meiosis triplets of consecutive informative markers are used: for a particular marker locus the FlexQTLTM software screens the closest left and right flanking informative meiosis (Figure ESM3.4). The number of observed double recombination events is calculated from the marker data and their order given the marker map. The number of expected double recombination events is calculated from the given map distances, assuming Haldane’s mapping function (Haldane 1919). Discrepancies indicate erroneous map positions of marker-loci or major errors in scoring. Following this check, four SSR loci had to be re-allocated to different chromosomes, two because of simple administrative errors and two due to inadequate assignment of the loci from a two-locus SSR. Figure ESM3.3: Fraction of informative meiosis events for all 87 SSR loci on 17 chromosomes. Figure ESM3.4: Fraction of observed (red symbols) and expected (black symbols) double recombinants for all 87 SSR loci. Upward (downward) triangles refer to the meiotic events between offspring and 1st (2nd) parent. References Evans KM, Patocchi A, Rezzonico F, Mathis F, Durel CE, Fernández-Fernández F, Boudichevskaia A, Dunemann F, Stankiewicz-Kosyl M, Gianfranceschi L, Komjanc M, Lateur M, Madduri M, Noordijk Y, Van de Weg WE (2010) Genotyping of pedigreed apple breeding material with a genome covering set of SSRs: Trueness to type of cultivars and their parentages. Mol Breeding 28:535-547 doi 10.1007/s11032-010-9502-5 Haldane JBS (1919) The combination of linkage values and the calculation of distances between the loci of linked factors. Journal of Genetics 8:299-309 Silfverberg-Dilworth E, Matasci CL, Van de Weg WE, Van Kaauwen MPW, Walser M, Kodde LP, Soglio V, Gianfranceschi L, Durel CE, Costa F, Yamamoto T, Koller B, Gessler C, Patocchi A (2006) Microsatellite markers spanning the apple (Malus x domestica Borkh.) genome. Tree Gen Genom 2:202224 Van Dijk T, Noordijk Y, Dubos T, Visser RGF, Van de Weg WE (2012) Microsatellite Allele Dose and Configuration Establishment (MADCE): An integrated approach to genetic studies in allopolyploids. BMC Pl Biol 12: 25 Voorrips RE (2002) MapChart: Software for the graphical presentation of linkage maps and QTLs. J Heredity 93:77-78