2014-05-21_12.15.09_d
advertisement

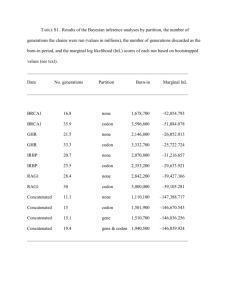
Journal of Nano Chemical Agriculture , Vol(1) , No(3) 80 PH Effects On 273th Codon Of P53 Gene By Computational Methods Nastaran Asghari Moghaddam1 Department of Biology, College of Basic Science, Tehran Science and Research Branch, Islamic Azad University, Tehran, Iran namoghadam@gmail.com Abstract P53 tumor suppressor gene, also known as “genome guardian” is mutated in more than half of all kind of cancers. In this study we have investigated the controls of environmental pH for P53 gene mutation in point of specific sequence which is prone to mutagenesis. The most probable cancerous mutations occur as point mutations in exons 5 -8 of P53 gene. By experimental research, it is revealed that acidic pH raised the rate of cancer and mutation in 273CGT codon of P53 gene. It to some extent is due to protonation of this three nucleotide codon. Mutation in this codon changes the encoding amino acid and subsequently produces a protein which has oncogenic features instead of tumor suppressor characteristics of original p53 protein. In current study, we perform investigation on the impact of protonation on stability of codon 273CGT in this gene. We used HYPERCHEM software for answering to our mentioned goal above.Our results suggested a reliable answer about the effect of protonation on mentioned codon and its stability. From theoretical point of view, acidity can increase the unstability of this specific codon. Along with the experimental investigations, our results can to some extent elaborate the mutagenesis of acidic pH. Keywords: P53 gene, protonation, mutagenesis Introduction P53 is a tumor-suppressor protein which has classical features of transcription factor. It responses to different cellular stress via inducing activation or repression of more than 2500 genes [2]. Because of its critical role in protection against cancer, it is called “the guardian of genome”. Somatic mutation of this gene has been observed in more than 50 percent of different human cancers [16]. Unlike other tumor suppressor genes (like RB, APC and BRCA1) in which inactivation occurs by frameshift and nonsense mutations, about 80 percent of P53 mutations are missense [7]. In P53 gene the most prevalent site of mutagenesis is along 5-8 exons, which encodes DNA binding domain. For instance, codon 273 (with 3 nucleotide sequence of CGT) included 8.7 percent of all mutations in P53 gene [9]. The importance of studying mutational pattern is to better understand the function of p53 domains, their effects on tumor- suppressing properties of p53 and the nature of etiologic substances as environmental etiologic biomarkers [13]. In addition, observations suggest that the pH of tumor microenvironment is more acidic than normal cells [4]. It is shown that disease “Barrett’s esophagus” is related to acid reflux [15] and even in its early stage mutation in P53 has been observed [5]. 81 PH Effects On 273th Codon Of P53 Gene Although the role of acidic pH is significant in carcinogenesis, its molecular mechanism is little known. It is known that physical properties of DNA is crucial for molecular genetic studies [10]. DNA molecule is constantly exposed to a great range of physical and chemical substances which harm its structure [14]. The effect of pH on DNA structure is not fully evaluated, because of the difficulty of measuring this quantity in cellular nucleus. In this regard, simulation and computational chemistry could be helpful. In current study, we tried to evaluate a new carcinogenesis pathway caused by pH alteration and creates missense mutation. We specifically focused on 273 codon (figure 1) of P53 gene. Materials and Methods CGT three-nucleotide was drawn as a double stranded DNA structure by HyperChem™. Then the structure was optimized by geometric optimization order. To determine the effect of pH on this structure a periodic box with 30 °A dimensions was designed. In addition, according to pH corresponding H+ and OH- for pH values 6.8 and 7.4 were respectively located within a periodic box. Simulation was done in MM+, AMBER, BIO+ and OPLS force fields. Molecular Mechanics calculations were assessed by Monte Carlo method [12]. Three important energy parameters – kinetic energy, potential energy and total energy- in four different simulating temperatures (308, 310, 312 and 314 Kelvin) were used for computation. Results and Discussion More than half of all human cancers have mutation in P53 gene. Theoretically evaluation of environmental factors like pH can help us to understand the causes of mutation and its molecular mechanisms. In current study computations were done in sophisticated and appropriate molecular modeling environment of HyperChem™ which is well known for its quality and flexibility [6, 11]. It is known that atoms are held together by forces. Function of biological systems arises from interaction of resilient bonds between atoms and electron motion. The main purpose is to seek for the lowest energy, in which the molecule is in its most stable state [8, 17]. In this study AMBER, MM+, BIO+ and OPLS force fields were chosen. When CGT is modeled, it undergoes shaking, rotating, stretching, and etc. functions around its bonds. The total potential energy is the sum of mentioned contribution interactions based on the force fields. Therefore, force fields are a series of functional energy parameters that evaluate performance and calculate the potential energy of molecule in various positions of its constituent atoms and bonds [3]. MM+ is a proper parameter for attaining vibration motion of atoms, related bond stretching potential, and angles bending. AMBER force field has extensive application for proteins and nucleic acids. It assigns all conformational energies and treats with hydrogen bond energy, and torsion term [1]. Like AMBER, OPLS is designed for computation of proteins and nucleic acids. In this force field bonded potentials are similar to AMBER and its non-bonded potentials involves vander waals and electrostatics. BIO+ filed is an extended form of CHARMM. Similar to AMBER and OPLS it has been designed to study macromolecules [8]. CGT codon was simulated in mentioned force fields in 4 different temperature (308K, 310K, 312K and 314K). To elucidate the effect of CGT energy on molecular mechanic calculation, the most usual expression for total potential energy is given by the following equation: 82 Journal of Nano Chemical Agriculture , Vol(1) , No(3) 𝑏𝑜𝑛𝑑𝑠 𝐸 𝑡𝑜𝑡𝑎𝑙 = ∑ 𝐸𝑖𝑠𝑡𝑟𝑒𝑡𝑐ℎ 𝑖 𝑏𝑜𝑛𝑑𝑎𝑛𝑔𝑙𝑒𝑠 + ∑ 𝐸𝑖𝑏𝑒𝑛𝑑 𝑖 𝑑𝑖ℎ𝑒𝑑𝑟𝑎𝑙𝑎𝑛𝑔𝑙𝑒𝑠 + ∑ 𝑎𝑡𝑜𝑚𝑝𝑎𝑖𝑟𝑠 𝐸𝑖𝑡𝑜𝑟𝑠𝑖𝑜𝑛 + ∑ 𝑖 𝑖𝑗 Covalent Interactions 𝐸 𝑣𝑎𝑛𝑑𝑒𝑟𝑤𝑎𝑎𝑙𝑠 + ∑ 𝐸 𝑒𝑙𝑒𝑐𝑡𝑟𝑜𝑠𝑡𝑎𝑡𝑖𝑐𝑠 𝑖𝑗 Non- Covalent Interactions E total is the sum of bonded and non-bonded interactions Ebond is stretching bond energy between two atoms Eangle is energy of bending an angle Etorsion is torsion energy of rotation around a bond E electrostatic and Evander waals are two energies which are exponent distribution, and repulsion or attraction between non-bonded atoms, respectively. The other two calculated energy quantities are kinetic and total energy values. In symbols the total energy equals: 𝐸𝑡𝑜𝑡𝑎𝑙 = ∑ 𝐸𝑝𝑜𝑡𝑒𝑛𝑡𝑖𝑎𝑙 + ∑ 𝐸𝑘𝑖𝑛𝑒𝑡𝑖𝑐 𝑎𝑡𝑜𝑚𝑝𝑎𝑖𝑟𝑠 (2) From a statistical point of view, the obtained valuable data for three basis sets of thermodynamic parameters (Ekinetic, Epotential, Etotal), analyzed under the different simulation procedure, various temperatures and different pH values every 10 ps span are listed in tables1, 2 and 3. According to results observed in table 1, the amount of kinetic energy constantly increase as the temperature was been raised. Obtained figures for kinetic energy in different time steps and various force fields were constant and the maximum quantity observed in314 K, 177.83 and (1) 213.4 Kcal/mol for pH values of 7.4 and 6.8, respectively. In acidic condition the amount of kinetic energy was approximately 25 percent more than physiologic pH. It is known that to have optimum function in biologic system, the energy levels must be in the minimum level. By comparing data from different pH values (see table 2), it is clear that potential energy of acidic pH is higher than physiologic pH and this increase can be observed in different force fields. In figure 2 minimum potential energy calculated by AMBER force filed in pH 7.4 and 6.8 have been reported. The increase of potential energy can be observed in this graph. It is notable that molecular stability in physiologic pH is more. Minimum potential energy levels in normal body temperature were 1918.23 and 289.57 for pH 6.8 and 7.4, respectively. Data analysis of table 3 exhibited that total energy quantities were affected by reducing pH value and increasing temperature. Current study was done to evaluate the thermodynamic role of acidic pH in creation of mutation in 273th codon of P53 gene. Analysis of kinetic, potential and total energies in 4 molecular mechanic energy fields. Increase of energy level has been observed in all three mentioned 83 PH Effects On 273th Codon Of P53 Gene parameters. Energy increase leads to molecular instability. According to the obtained results in this study, it can be concluded that acidic condition in cellular environment directly produce unstable codon structure which can cause changes in codon structure. If this alteration in biologic system is not recognized and not repaired, it can result in the mutation of DNA. Regarding to the fact that genetic changes’ negative role in cells, this can be a beginning point for cancer progression. The results can be a cancer with a poor prognosis if this alteration happens in a gene like P53. References 1. Case, D.A., Cheatham, T.E. III, Darden, T., Gohlke, H., Luo, R.,Merz, K.M., Onufriev, A., Simmerling, C., Wang, B., Woods, R., 2005. The Amber biomolecular simulation programs.J.Comput. Chem., 26, 1668–1688. 2. Cui, F., Sirotin, M.V., Zhurkin, V.B., 2011. Impact of Alu repeats on the evolution of human p53 binding sites, Biol. Direct., 6: 2. 3. Foloppe, N., Mackerell, A.D., 2000. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem., 21:86-104. 4. Helmlinger, G., Yuan, F., Dellian, M., Jain, R.K., 1997. Interstitial pH and pO2 gradients in solid tumors in vivo: high-resolution measurements reveal a lack of correlation, Nat. Med., 3: 177-182. 5. Keswani R.N., Noffsinger, A., Waxman, I., Bissonnette, M., 2006. Clinical use of p53 in Barrett's esophagus, Cancer Epidemiol. Biomarkers Prev., 15(7):1243-9. 6. Lewars, E., 2003. Introduction to the theory and application of molecular and quantum mechanics, J. Computational chemistry, Ontario Canada 7. Li, F.P., Fraumeni, J.F., Mulvihill, J.J., Blattner, W.A., Dreyfus, M.G., Tucker, M.A., Miller, R.W., 1988. A cancer family syndrome in twenty-four kindreds, Cancer Res., 48(18): 5358–5362. 8. Mackerell, A. D., 2004. Empirical force fields for biological macromolecules: Overview and issues, J. Comput. Chem, 25: 1584–1604. 9. Mogi, A., Kuwano, H., 2011. TP53 mutations in nonsmall cell lung cancer, J Biomed Biotechnol, 583929: 9 pages. 10. Monteiro Carlos, E.T.B., 2008. Acidic environment evoked by chronic stress: A novel mechanism to explain atherogenesis. Available from Infarct Combat Project at http://www.infarctcombat.org/AcidityTheory .pdf 11. Nam, K.,Gao, J., York, D. M., 2008.Quantum mechanical / molecular mechanical simulation study of the mechanism of hairpain ribozyme catalysis, J. Am. Chem. Soc. ,130: 4680-4691. 12. Norberg, J., Nilsson, L., 2002. Molecular dynamics applied to nucleic acids, AcChem Res, 35(6): 465-72. 13. Peltonen J.K., Helppi, H.M., Pääkkö, P., Turpeenniemi-Hujanen, T., Vähäkangas, K.H., 2010. P53 in head and neck cancer: functional consequences and environmental implications of TP53 mutations, Head Neck Oncol, 2:36. 14. Pinchuk, A., 2004. Optical constants and dielectric function of DNA’s nucleotides in UV range. Journal of Quantitative Spectroscopy & Radiative Transfer, 85: 211–215. 15. Prescott, D.M., Charles, H.C., Poulson, J.M., Page, R.L., Thrall, D.E., Vujaskovic, Z., Dewhirst, M.W., 2000. The relationship between intracellular and extracellular pH in spontaneous canine tumors, Clin Cancer Res., 6(6): 2501-5. 84 Journal of Nano Chemical Agriculture , Vol(1) , No(3) Weiner, P.K., 1984. A new force field for molecular mechanical simulation of nucleic acids and proteins, J. Am. Chem. Soc., 106: 765-784. 16. Vogelstein, B., Lane D., Levine, A.J., 2000. Surfing the P53 network, Nature, 408: 307310 17. Weiner, S.J., Kollman, P.A., Case, D.A., Singh, U.C., Ghio, C., Alagona, G., Profeta, S., Table 1. Computed CGT Kinetic energy ( kcal/ mol ), belong to AMBER, MM+, BIO+ and OPLS force fields under four different temperature and 2 various pH values. Kinetic Energy (K Cal/mol) 6.8 pH condition 7.4 Method 308 K 310 K 312 K 314 K 308 K 310 K 312 K 314 K AMBER 209.3228 210.6821 212.0413 213.4005 174.4357 175.5684 176.7011 177.8338 BIO + 209.3228 210.6821 212.0413 213.4005 174.4357 175.5684 176.7011 177.8338 MM + 209.3228 210.6821 212.0413 213.4005 174.4357 175.5684 176.7011 177.8338 OPLS 209.3228 210.6821 212.0413 213.4005 174.4357 175.5684 176.7011 177.8338 Table 2. Computed CGT potential energy ( kcal/ mol ), belong to AMBER, MM+, BIO+ and OPLS force fields under four different temperature and 2 various pH values. Potential Energy (Kcal/mol) Amber/Monte Carlo 6.8 Method pH condition time (PS) 308 K 310 K 10 20 30 40 50 60 70 80 90 100 42186.5 10721.1 6389.2 4700.9 3792.5 3159.3 2751.5 2432.0 2154.7 1955.1 308 K Method pH condition time (PS) 10 20 30 40 50 60 70 80 90 100 310 K 312 K MM+/ Monte Carlo 7.4 314 K 308 K 310 K 312 K 314 K 308 K 39561.2 30530.5 9311.3 8789.2 5942.6 5670.5 4604.6 4428.1 3764.8 3643.5 3172.1 3035.1 2761.0 2625.5 2447.3 2307.0 2198.2 2098.2 2002.8 1928.2 BIO/Monte Carlo 6.8 33473.4 9425.3 6103.5 4699.8 3861.3 3318.5 2875.8 2533.6 2246.3 2055.2 781.4 515.8 416.0 389.3 359.7 339.7 327.6 309.2 297.0 303.0 805.8 783.7 522.4 522.9 398.9 447.6 354.7 396.9 337.0 371.4 338.5 340.6 326.8 322.6 321.2 311.9 319.7 310.0 303.2 292.9 BIO/Monte Carlo 7.4 822.3 559.7 448.1 396.5 371.0 342.9 334.6 332.4 322.5 325.8 11706.2 7401.4 5039.8 3822.3 3150.9 2632.8 2258.5 2027.6 1785.0 1625.2 310 K 312 K 314 K 308 K 310 K 312 K 314 K 308 K 310 K 312 K 314 K 308 K 310 K 312 K 314 K 58710.0 11105.1 6354.2 4815.2 3868.6 3262.4 2857.5 2545.7 2303.6 2133.3 50746.8 10265.9 6578.6 4881.7 3893.0 3272.0 2844.3 2508.5 2256.5 2048.8 1224.7 801.8 675.4 628.9 585.8 550.1 540.1 536.7 520.9 503.3 1228.0 777.6 684.7 639.3 608.5 575.9 560.5 547.7 533.6 517.1 1186.7 844.4 690.5 621.6 597.2 568.5 560.5 554.0 531.8 526.7 1266.6 882.0 725.5 663.3 621.1 588.4 553.9 534.9 525.5 520.5 29556.5 10776.5 7889.3 6743.5 6010.4 5514.9 5134.9 4854.6 4644.2 4415.4 25226.0 10091.2 7687.4 6593.8 5860.1 5355.9 5047.3 4776.0 4568.5 4407.5 26018.3 10623.3 7791.8 6596.0 5894.0 5393.2 5022.1 4787.9 4581.8 4446.9 28227.2 10002.7 7482.3 6491.0 5785.9 5335.7 4994.3 4775.5 4579.7 4429.9 823.8 506.2 430.8 405.3 378.3 358.8 326.7 305.5 299.8 298.3 765.4 495.4 406.9 368.9 369.4 343.5 317.8 306.3 290.2 282.1 868.8 527.5 435.6 371.8 355.4 337.8 320.9 324.8 309.4 307.2 815.6 509.3 427.1 375.9 348.8 341.2 327.2 323.8 309.3 305.2 112634.2 42848.9 25535.1 9631.3 10619.4 6449.1 6056.7 4973.2 4452.3 4024.0 3796.8 3358.4 3202.9 2881.6 2807.6 2523.1 2509.9 2248.1 2285.2 2016.9 312 K MM+/ Monte Carlo 6.8 Amber/Monte Carlo 7.4 314 K 11573.7 11747.1 11725.9 7501.0 7244.9 7489.7 5306.3 5088.0 5190.4 4002.1 3772.1 3995.4 3228.8 3028.1 3185.2 2697.7 2517.7 2655.3 2310.3 2137.8 2310.6 2023.6 1901.9 2012.6 1796.8 1689.1 1784.9 1610.9 1524.3 1633.3 OPLS/ Monte Carlo 6.8 308 K 1071.6 730.7 630.4 569.2 527.3 500.6 501.6 498.9 487.8 474.1 310 K 312 K 1100.3 1156.1 715.5 756.5 614.6 651.5 569.9 585.1 544.1 557.7 518.3 526.9 489.5 517.3 488.0 505.7 472.9 496.5 474.1 485.8 OPLS/ Monte Carlo 7.4 314 K 1094.2 744.6 610.2 566.6 535.8 521.5 514.7 503.7 494.7 468.6 PH Effects On 273th Codon Of P53 Gene 85 Table 3. Computed CGT total energy ( kcal/ mol ), belong to AMBER, MM+, BIO+ and OPLS force fields under four different temperature and 2 various pH values. Total Energy (Kcal/mol) Amber/Monte Carlo 6.8 Method pH condition time (PS) 308 K 310 K 312 K 314 K 42395.83 39771.92 30742.54 33686.84 10930.47 9521.98 9001.24 9638.72 6598.47 6153.27 5882.54 6316.91 4910.22 4815.23 4640.13 4913.24 4001.78 3975.48 3855.50 4074.67 3368.65 3382.83 3247.16 3531.85 2960.81 2971.70 2837.54 3089.22 2641.34 2657.94 2519.08 2747.03 2364.04 2408.91 2310.24 2459.74 2164.41 2213.48 2140.25 2268.61 Method BIO/Monte Carlo pH condition 6.8 10 20 30 40 50 60 70 80 90 100 time (PS) 10 20 30 40 50 60 70 80 90 100 308 K 310 K 312 K MM+/ Monte Carlo 6.8 Amber/Monte Carlo 7.4 314 K 112843.51 43059.53 58922.06 50960.23 25744.46 9842.00 11317.15 10479.31 10828.69 6659.75 6566.27 6791.99 6266.05 5183.86 5027.21 5095.12 4661.60 4234.73 4080.61 4106.44 4006.14 3569.05 3474.48 3485.43 3412.18 3092.25 3069.52 3057.70 3016.90 2733.80 2757.75 2721.93 2719.21 2458.83 2515.61 2469.92 2494.55 2227.63 2345.34 2262.16 308 K 955.79 690.26 590.43 563.78 534.09 514.13 502.07 483.65 471.40 477.48 310 K 312 K 314 K 308 K 310 K 312 K MM+/ Monte Carlo 7.4 314 K 308 K 981.37 960.40 697.96 699.62 574.51 624.35 530.22 573.58 512.58 548.12 514.11 517.25 502.41 499.34 496.77 488.61 495.29 486.75 478.73 469.58 BIO/Monte Carlo 7.4 1000.10 11915.49 11784.35 11959.10 11939.31 737.49 7610.70 7711.67 7456.99 7703.11 625.93 5249.14 5517.01 5300.08 5403.80 574.36 4031.59 4212.79 3984.11 4208.78 548.84 3360.27 3439.43 3240.12 3398.65 520.73 2842.07 2908.40 2729.76 2868.73 512.44 2467.79 2520.97 2349.84 2523.96 510.17 2236.90 2234.25 2113.97 2225.97 500.31 1994.32 2007.45 1901.19 1998.26 503.66 1834.50 1821.63 1736.35 1846.69 OPLS/ Monte Carlo 6.8 1246.02 905.11 804.79 743.64 701.73 675.08 676.04 673.34 662.26 648.49 308 K 310 K 312 K 314 K 308 K 1399.09 976.22 849.85 803.33 760.25 724.57 714.50 711.11 695.29 677.76 1403.60 953.17 860.25 814.83 784.11 751.48 736.11 723.29 709.13 692.66 1363.36 1021.14 867.17 798.34 773.92 745.20 737.18 730.70 708.46 703.36 1444.39 29765.77 25436.72 26230.32 28440.65 1059.83 10985.77 10301.88 10835.39 10216.09 903.29 8098.60 7898.10 8003.80 7695.71 841.08 6952.81 6804.52 6808.02 6704.35 798.89 6219.72 6070.74 6106.00 5999.28 766.19 5724.22 5566.56 5605.20 5549.11 731.74 5344.17 5257.97 5234.13 5207.72 712.71 5063.88 4986.71 4999.95 4988.89 703.30 4853.53 4779.15 4793.89 4793.11 698.32 4624.72 4618.22 4658.94 4643.34 308 K 310 K 312 K 314 K Figure 1. Molecular structure of CGT three-nucleotide 998.19 680.60 605.19 579.71 552.70 533.25 501.11 479.96 474.26 472.73 310 K 312 K 1275.82 1332.79 891.08 933.21 790.17 828.23 745.49 761.84 719.70 734.36 693.85 703.59 665.04 693.98 663.57 682.38 648.47 673.22 649.62 662.48 OPLS/ Monte Carlo 7.4 310 K 940.98 670.98 582.44 544.45 544.94 519.12 493.40 481.92 465.81 457.67 312 K 1045.46 704.19 612.29 548.54 532.13 514.48 497.62 501.50 486.14 483.85 314 K 1272.06 922.44 787.98 744.48 713.61 699.31 692.58 681.55 672.53 646.48 314 K 993.43 687.15 604.92 553.75 526.68 519.06 505.06 501.65 487.14 483.06 Journal of Nano Chemical Agriculture , Vol(1) , No(3) Figure 2. Comparison of minimum energy level in different temperatures in AMBER force field 86