We have made corrections for manuscript "Piper betle induces
advertisement

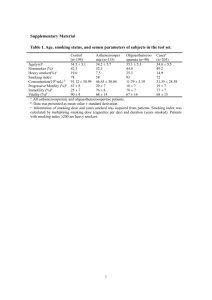
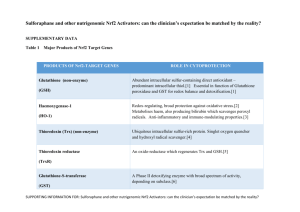
We have made corrections for manuscript "Piper betle induces phase II genes and proteins through Nrf2/ARE signaling pathway in mouse embryonic fibroblasts cells in culture" according to comments made by reviewers 1, 2 & 3.Answers given are in blue text. REVIEWER 1 (Ki Churl Chang): • In figure 2, it is questionable whether detection of Nrf2 after 10, 12 h treatment with PB in Nrf2KO cells, because no Nrf2mRNA detected in Nrf2KO cells in fig4a. This can be explained by the time point at which the mRNA was measured. We measured mRNA by qRT-PCR technique, and by 24 hr perhaps mRNA has all been transcribed to Nrf2 protein, and thus they may not be detected. • It needs explanation for Fig4 where it decreases HO-1 mRNA but increased protein level. We have added the following explanation in the manuscript (highlighted yellow in the manuscript). ‘Our results showed decreased HO-1 mRNA expression after 24 h treatment with PB while its protein level was found to be increased. This finding is similar to a time-course study by Motterlini et al. [42] who showed that HO-1 mRNA expression in bovine aortic endothelial cells reached a maximum after 6 h treatment with 5 μM curcumin and declined after 24 h treatment.’ • In figure 5, it seems like that greater increase of HO-1 by PB is in WT than NO. Yes, it is true that PB increased HO-1 protein in WT cells compared to N0 cells (Fig. 5). REVIEWER 2 (Jeff Fry): 1. You can’t have “transcriptional induction of...GSH”, although as stated later you can have transcriptional induction of the enzymes responsible for the synthesis of GSH. Reword. We have made corrections as suggested (highlighted yellow in the manuscript). ‘This leads to transcriptional induction of a number of antioxidant enzymes responsible for the synthesis of heme oxygenase-1 (HO-1), superoxide dismutase (SOD), catalase (CAT), phase I oxidoreductase [NAD(P)H quinone oxidoreductase (NQO1)], phase II detoxifying enzymes [glutathione S-transferase (GST)], and including glutathione biosynthesis enzymes glutathione cysteine ligase modifier subunit (GCLM) and glutathione cysteine ligase catalytic subunit (GCLC) [15-17].’ 2. Indicate the concentrations of penicillin-streptomycin and amphotericin-B used in the culture medium We have added the concentrations of penicilin-streptomycin and amphotericin-B in the manuscript. ‘MEFs cells were cultured in Iscove’s modified Dulbecco’s medium (Invitrogen, USA) supplemented with 10% FBS (PAA, USA), 10 unit/ml penicillin-streptomycin and 2.5 μg/ml amphoterecin-B (PAA, USA).’ 3. Indicate the post-hoc test used after the ANOVA. Differences between mean values of multiple groups were analyzed by one-way analysis of variance (ANOVA) with Tukey’s HSD post-hoc test. 4. P9, line 3 – Remove “it” (also P11, line 6 and line 16) We have already removed ‘it’ from the pages stated. 5. P9, 3 lines from bottom – “significantly expressed” is very clumsy. What I assume is meant is that, at the basal state, the level of expression of the SOD gene is significantly greater in N0 cells compared to WT cells. Reword as appropriate. We have changed accordingly as suggested to read, “ SOD1 gene expression is significantly greater in N0 cells compared to WT cells at the basal state.” 6. P10, line 5 of Discussion – Reword as “The study illustrated that Nrf2 deficiency in MEF cells...” We have reworded as suggested 7. P10, line 9 of Discussion – [20] not (20) This is corrected as suggested. 8. P10, last 7 lines – Clumsy expression. Reword to clarify meaning. As suggested by 3rd reviewer, we decided to use another reference. According to the 3rd reviewer, it seems inappropriate to compare PB extract to the known hepatotoxicant, as it implies PB extract may be a hepatotoxicant. It now reads as below: ‘This study illustrated that Nrf2 deficiency in MEF cells (N0) resulted in increased cytotoxicity with PB treatment compared to WT cells. Hydroxychavicol which is found in PB was shown to exert antioxidant activity at low concentration but display pro-oxidant effect at concentration higher than 0.1 mM [22]. A well-known Nrf2 activator and antioxidant, epigallocatechin 3-gallate (EGCG) which inhibits H1299 human lung cancer at 0-35 uM was shown to be pro oxidant at higher concentration (50 uM) by inducing the production of intracellular ROS in human lung cancer cells [23].’ 9. P12, line 17 – Reword as “proximal cells.” This is corrected as suggested. 10. P13, line 5 – Replace “it is” with “were”. We have replaced “it is” with “were” as suggested (highlighted yellow in the manuscript). 11. P21, legend to Figure 1 – The figure itself shows no asterisks, so replace the statement in the legend. The legend has been corrected, as suggested. 12. Figure 1 – Express the x-axis values on a true linear scale. We have already expressed the x-axis values on a true linear scale. REVIEWER 3 (Scott A Reisman): Major Comments: 1. The article would be enhanced with better grammatical proofreading and subject-verb agreement and comma placement corrections, etc. One of the authors WZWN who is well versed in English had revised the paper to ensure there were no grammatical errors. 2. Newer data and models support that Nrf2 does not “dissociate” from Keap1. This should be corrected and in-line with more recent literature. The text throughout the manuscript should be updated to reflect this more recent knowledge. We have included in the manuscript recent knowledge on Nrf2 as suggested. It is now part of the Introduction. ‘Nrf2 is normally sequestered in the cytoplasm as an inactive complex by interaction with its repressor Keap1 [10]. There are two prevailing models which proposed how Nrf2 is released from Keap1 in response to oxidative stress; (1) the ‘hinge and latch’ model, in which Keap1 modifications in thiol residues of Keap1 disrupt the interaction with Nrf2 causing misalignment of the lysine residues within Nrf2 that can no longer be polyubiquitinylated, and (2) the model in which thiol modification causes dissociation of Cul3 from Keap1 [11]. In both models, the inducer-modified and Nrf2-bound Keap1 is inactivated and, consequently, newly synthesized Nrf2 proteins bypass Keap1 and translocate into nucleus, bind to the Antioxidant Response Element (ARE) and leads to transcriptional induction of Nrf2 target genes such as heme oxygenase-1 (HO-1), superoxide dismutase (SOD1), catalase (CAT), NAD(P)H quinone oxidoreductase (NQO1), glutathione S-transferase (GST), and including glutathione biosynthesis enzymes glutathione cysteine ligase modifier subunit (GCLM) and glutathione cysteine ligase catalytic subunit (GCLC) [11-17]. 3. The last sentence of the introduction should be expanded with greater detail added. The type of “knockout cells” should be described here. We have added the text below at the end of introduction section. ‘The aim of this experiment was to evaluate whether PB can modulate Nrf2/ARE pathway with subsequent increase in phase 2 and endogenous antioxidant enzymes using mouse embryonic fibroblast (MEF) from wild-type and Nrf2 knockout mice. Here, we report that PB treatment stimulates the Nrf2 pathway which leads to elevation of multiple Nrf2-regulated genes in wild-type (WT) fibroblast but not in Nrf2 deficient cells (N0) . 4. I do not agree that the Nrf2 level of translocation shown in Figure 2 by PB is comparable to SFN. SFN appears markedly greater. We agree with the reviewer regarding this matter and as such we have changed accordingly as below. ‘As compared to wild-type cells treated with 5 µM sulforaphane for 6 h that serves as the positive control, our time-course study showed that treatment with PB for 10 h activates Nrf2 and translocates it into the nucleus at a lower level compared to its activation by SFN.’ 5. The mRNA and protein Nrf2 target protein data do not all together clearly demonstrate Nrf2 activation, particularly for HO-1 and SOD1, which are not always the best for evaluation of Nrf2 engagement. It would be better to show more targets which demonstrate a dosedependent increase in WT but not Nrf2 KO cells. Other better Nrf2 targets should be selected and evaluated. This reviewer suggests Gclc, Gclm, a Gst, Txnrd1, and/or Srxn1. We have tried to include the genes GCLC and GCLM as suggested by the reviewer in one of our earlier experiments, however, we failed to get any good results even though we have attempted several times. Thus we decided to abort using these genes. 7. The manuscript may benefit from the use of 3 concentrations of PB extract instead of 2. Based on the viability assay (Figure 1), we have decided to choose 2 concentrations (5 and 10 μg/ml) as concentrations higher than 10 μg/ml can inhibit the growth of the knockout cells which are more sensitive to WT. Minor Comments: 1. Nqo1 is not a phase II enzyme (e.g. Gsts, Ugts, and Sults) but a phase I oxidoreductase enzyme. This is a common error in the literature and should be corrected throughout the manuscript. It has been already corrected throughout the manuscript that Nqo1 is a phase I oxidoreductase, and not a phase II enzyme. 2. In the introduction, please elaborate on the following sentence. “Aqueous extract of PB leaves prevents lipid peroxidation better than tea leaves extract using in vitro model.” What in vitro model? We have removed this reference and replaced with a new reference. It now reads as below: ‘Aqueous extract of PB leaves provides better protection against tissue lipid peroxidation in diabetic rats’ 3. This sentence needs a reference: “Oxidative stress results from an imbalance between excess production of ROS and limited cellular antioxidant defense.” The reference below is added in the manuscript. ‘Sies H: Oxidative stress: Oxidants and Antioxidants. Experimental Physiology 1997, 82, 291-295.’ 4. The mouse strain background of the cells should be provided. Please provide a reference, if possible. Mouse embryonic fibroblasts (MEFs) were prepared from wild type (Nrf2+/+ ) and nrf2disrupted (Nrf2 -/- ) ICR mice and they were a kind gift from Kwak Mi-kyoung (The Catholic University of Korea, Bucheon, South Korea) and Nobunao Wakabayashi (University of Pittsburgh, USA). This is usually how it is described in the papers by Kwak Mi-Kyoung: M.K. Kwak, N. Wakabayashi, J.L. Greenlaw, M. Yamamoto, TW. Kensler. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway, Mol.Cell. Biol. 23 (2003) 8786-8794 5. Concentrations of PB extract used to treat cells should be mentioned often in the methods section. We have added accordingly where necessary the concentrations of PB extract used to treat MEF cells, 5 and 10 μg/ml throughout the method section. 6. What is “lysis buffer?” Lysis buffer consists of a cocktail containing RIPA buffer and (Sigma, USA) and a protease inhibitor cocktail tablet (Roche, Switzerland). This sentence has been added in the method section. 7. Please use “x g” instead of “rpm.” We have replaced ‘rpm’ with ‘x g’, as suggested. 8. What is “Nrf2 dislocation?” We have changed ‘Nrf2 dislocation’ to ‘Nrf2 nuclear level’. ‘After 10 h of 10 µg/ml PB treatment, there was a significant difference (p<0.05) of Nrf2 nuclear level in WT cells compared to N0 cells.’ 9. Why does Nrf2 protein appear to be detected in the knockout cells? This maybe due to the antibody that we employed in this study. Antibodies from various companies have been shown to vary in their specific binding to the respective antigen. However, conformation by real time PCR showed that mouse embryonic fibroblasts of Nrf2 Nrf2 knockout cells showed no expression of Nrf2 gene . 10. The legend for Figure 2 does not match the figure. Should there be a Figure 2D? Please indicate the time points in the bar graphs. We have indicated the time points in the bar graphs, as suggested. 11. Please provide a reference for the Nqo1-ARE sequence used for the luciferase assay. We received the plasmid Nqo1-ARE for luciferase assay from Nobunao Wakabayashi (University of Pittsburg, USA) and Kwak Mi-Kyoung (The Catholic University, Bucheon, South Korea). Previous studies which used the same plasmid are listed below: 1 Sarala Manandhar, Jeong-Min Cho, Jung-Ae Kim, Thomas W. Kensler, Mi-Kyoung Kwak. Induction of Nrf2-regulated genes by 3H-1, 2-dithiole-3-thione through the ERK signaling pathway in murine keratinocytes. European Journal of Pharmacology 2007, 577: 17–27 2 Sarala Manandhar, Aram You, Eung-Seok Lee, Jung-Ae Kim and Mi-Kyoung Kwak. Activation of the Nrf2-antioxidant system by a novel cyclooxygenase-2 inhibitor furan-2-yl-3-pyridin-2ylpropenone: implication in anti-inflammatory function by Nrf2 activator. Journal of Pharmacy and Pharmacology 2008, 60: 879–887 12. Please indicate the genes and proteins analyzed in the bar graphs in Figures 4 and 5, respectively. We have indicated the genes and proteins in the bar graphs for Figures 4 and 5, respectively. 13. I do not see any difference in HO-1 or SOD1 protein expression after PB treatment in the westerns in Figure 5. The differences between WT and KO are apparent. We have analyzed the Westerns band for NQO1, HO-1 and SOD1 using ImageMaster TotalLab v1.11 software and the values were plotted as Figure 5B, C and D. The data obtained represents for 6 bands with n = 3. After statistical analysis, we saw that HO-1 protein increased significantly in WT cells but not in N0 cells after PB treatment. However for SOD1, there was no significant difference after PB treatment. 14. It seems inappropriate to compare PB extract to the known hepatotoxicant diquat, as it implies PB extract may be a hepatotoxicant. We agree with the reviewer and have removed diquat as a reference to our finding. Instead we have added new reference to support our finding. It now reads as below in the text: ‘This study illustrated that Nrf2 deficiency in MEF cells (N0) resulted in increased cytotoxicity with PB treatment compared to WT cells. Hydroxychavicol which is found in PB was shown to exert antioxidant activity at low concentration but display pro-oxidant effect at concentration higher than 0.1 mM in oral KB carcinoma cells [22]. A well-known Nrf2 activator and antioxidant, epigallocatechin 3-gallate (EGCG) which inhibits H1299 human lung cancer at 0-35 uM was shown to be pro oxidant at higher concentration (50 uM) by inducing the production of intracellular ROS in human lung cancer cells [23].’