Introduction - Louisiana Tech University
advertisement

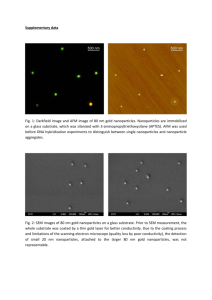
Nanodiamond Seeding of QCM for Biosensor Applications Student: Andre Schexnider Mentor: Adarsh Radadia Introduction Infections in United States from food-borne pathogens, Salmonella, E. coli, Campylobacter, and Listeria caused more than 3 billion dollars of economic damage in 2010 [1]. Food supplies must therefore be carefully monitored. One approach to such monitoring uses highly sensitive, selective, and stable sensors that can work continuously without constant removal and analysis. Quick detection is important. If the bacteria are detected early enough, the supply lines can be stopped immediately and the amount of infections will decrease. Biosensors that have been developed to detect bacteria have used antibodies, aptamers, and synthetic molecules to capture the bacteria. The captured bacteria can then detected by either the change in mass, electronic structure, or thickness of the capturing film [2]. Quartz Crystal Microbalances (QCM) can detect mass changes. Quartz crystals have a natural frequency, and frequency changes can be directly correlated to its mass [2]. However, the typical surfaces for QCM are gold, platinum, silicon dioxide, and silicon nitride, which can lead to unstable surfaces in liquid environment on prolonged exposure [4]. Radadia et al. have shown that ultrananocrystalline diamond (UNCD) surfaces have extended stability which makes this surface promising in the creation of longer lasting biosensors [3]. The aim of this project is to utilize the high stability of diamond and combine with the sensing ability of QCM, to create a real-time, highly selective biosensor that has a reasonable lifetime. Growth of UNCD is first accompanied by seeding of the surface in which small nanoparticles of diamond are sonicated on to the surface. These seeds are sites of nucleation. The assumption is that the seeding process will generate an evenly distributed surface of nanodiamond particles, where the antibodies would then be attached. Methods The sample was seeded with 3 concentrations of a mixture with ratios of 1:1, 1:3 and 1:5 slurry to methanol. The sonication time was varied at 5, 15, and 30 minutes. These samples were analyzed using AFM and SEM. Figure 1 shows the morphology of the diamond on the surface of the sample that was sonicated for 15 minutes in a 1:3 solution. Results and Conclusion The results demonstrated that the surfaces were coated with UNCD (Figure 1) and that the resulting surface roughness depended on sonication time and the ratio of diamond/DMSO slurry to methanol. 1 Figure 1: (Left) Scanning electron microscope image of a nodiamond-coated surface. (Right) Dependence of surface roughness on slurry content and sonication time. References [1] U. ERS. (2011). Foodborne Illness Cost Calculator. [2] M. Liss, B. Petersen, H. Wolf, and E. Prohaska, "An aptamer-based quartz crystal protein biosensor," Anal Chem, vol. 74, pp. 4488-95, 2002. [3] A. D. Radadia, C. J. Stavis, R. Carr, H. Zeng, W. P. King, J. A. Carlisle, et al., "Control of Nanoscale Environment to Improve Stability of Immobilized Proteins on Diamond Surfaces," Adv Funct Mater, vol. 21, pp. 1040-1050, 2011. 2 Effect of Particle Clustering and Geometry on Joule Heating in Therapeutic Nanoparticles Student: Nicholas K. Rader Mentor: Dentcho Genov Introduction: Gold nanoparticles, in the form of nanorods, exposed to 800 nm wavelength light, neutralize cancer cells without a biomarker [1]. In addition, immunotargeted nanoshells have been used to find and destroy breast carcinoma cells that overexpress HER2, a relevant cancer biomarker [2]. Gold nanoparticles have been applied to cancer imaging, spectroscopic detection, and photothermal therapy [3]. Surface plasmon resonance leads to intense electromagnetic fields on the particle surface that enhance absorption and scattering [3]. When gold nanoparticles undergo coupled surface plasmon resonance, they create strong electric fields which enhance imaging of cancerous cells and distinguish between malignant and nonmalignat cells [4]. Silver nanoparticles have also been studied in antiviral therapy, specifically, with the HIV-1 virus. The nanoparticles bind to the virus specifically in the spectrum of 1-10 nm making its interaction with the virus size-dependent [5]. Metal nanoparticles interact with each other uniquely. This unique interaction results in the exponential distance decay and the interesting “universal” scaling behavior of interparticle plasmon coupling [6]. It is possible to derive a “plasmon ruler equation” that estimates the interparticle seperation between gold nanospheres in a biological system from the observed fractional shift of the plasmon band. This equation allows metal nanoparticles to be optimized in chemical and biological imaging, sensing, and therapeutics. Methods In the present work, we used COMSOL to generate a geometric model of gold nanospheres and to solve Maxwell’s equations and Gauss’s laws to calculate surface plasmon resonance. From these simulations, we derived the resistive heating as a function of wavelength for circular spheres and elliptical spheres. Results Figure 2 displays the resistive heating as a function of wavelength for each spherical geometry, and Figure 3 shows the resisitive heating as a function of wavelength for the elliptical spheres. The combination of three spheres had the highest peak of 1.5 Joules at 360 nm, while one sphere had the second highest peak of 0.42 Joules at 390 nm. The largest heating for ellipsoids occurred with single particles (1.15 Joules at 4.27 nm), followed closely by four particles (0.93 Joules at 502 nm). 3 Figure 2: Resistive heating for different geometries of nanospheres. Red (one sphere), Green (two spheres), Blue (three spheres), Purple (four spheres). Figure 3: Red (one ellipsoid), Green (two ellipsoids), Blue (three ellipsoids), Purple (four ellipsoids) Conclusion Complex geomtries were as efficient at absorbing light through its surface plasmon resonance as simple geometries. However, the simpler geometries led to resistive heating that was more wavelength-specific. References [1] X. Huang, I.H. El-Sayed, W. Qian, and M.A. El-Sayed, “ Cancer Cell Imaging and Photothermal Therapy in the Near-Infrared Region by Using Gold Nanorods,” J. Am. Chem. Soc., vol. 128, pp. 2115-2120, 2006 4 [2] C.Loo, A. Lowery, N. Halas, J. West, and R. Drezek, “Immunotargeted Nanoshells for Integrated Cancer Imaging and Therapy,” Nano Lett., vol. 5, pp. 709-711, 2005 [3] X. Huang and M.A. El-Sayed, “Gold Nanoparticles: Optical properties and implementations in cancer diagnosis and photothermal therapy,” J. Advanced Research, vol. 1, pp.13-28, Jan. 2010 [4] J.C. Kah, K.W. Kho, C.G. Lee, C. James, R. Sheppard, Z.X. Shen, K.C. Soo, and M.C. Olivo, “Early diagnosis of oral cancer based on the surface plasmon resonance of gold nanoparticles,” Int. J. Nanomedicine, vol. 2, pp. 785-798, 2007 [5] J.L. Elechiguerra, J.L. Burt, J.R. Morones, A. Camacho-Bragado, X. Gao, H.H. Lara, and M.J. Yamacan, “Interaction of silver nanoparticles with HIV-1,” J. Nanobiotechnology, vol. 3, 2005 [6] P.K. Jain, W. Huang, and M. A. El-Sayed, “On the Universal Scaling Behavior of the Distance Decay of Plasmon Coupling in Metal Nanoparticles Pairs: A Plasmon Ruler Equation,” Nano Lett., vol. 7, pp. 2080-2088, 2007 5 Modeling glioma multicellular spheroids with novel particle-based growth generation by random walk model Student: Benjamin Cote Mentor: Mark DeCoster Introduction Glial cells are highly invasive, migratory cancer cells that follow well understood patterns of general cell migration (Frieboes 2007, Mehta 2012, Swanson 2003). Computational models that predict their behavior can be useful to fundamental research, diagnosis, and treatment. Computational models, based on first order diffusion equations, have been developed to predict the migratory behavior of these cells (Frieboes 2007). However, differences have been observed between the results of these models and the spheroid shapes that are obtained in vitro (Mehta 2012). An alternative method, random walk modeling (RWM) is applied in this study. The tumor shapes from this method are compared to those obtained through diffusion-based models and to those obtained in vitro. Methods The RWM program was implemented in MATLAB. A three-dimensional space is divided into small cubic volumes, where the height, width, and depth of each volume is one cell diameter. The program initializes each spheroid’s size and location, and growth rate data are used to determine the time for each cell division cycle. Each cell is considered to be in one of the small cubic volumes. At each cell division, a new cell is generated, and its position is randomly selected. A growth cycle runs through six directions in a bound matrix to track the spheroids in digital space and ensures that cells only grow once in a doubling through a second matrix. Three growth cycles occur in the program when any spheroid is requested. An initializing cycle places the single-celled locations of seeded spheroids. The monitored cycle collects data requested by the user on each doubling. A death cycle determines whether the nutrients available to the cell are low enough to cause cell death. Available nutrients are deduced from the distance of the cell to the culture medium. The desired number of cells was altered with each test. Measurements chosen for the experiment were determined by parameters that could be measured experimentally or from the literature, and one spheroid was set with each test. The measured outputs include the total number of cells, and the diameters and areas of the formed spheroids. Data extracted are the change in cell numbers with respect to doublings, change in cell growth, and the sphericity of each tumor. The negative control is experimentally formed tumors in vivo from literature, and the positive control is experimentally formed spheroids using previous models. ANOVA testing was used for statistical analysis. 6 Results Figure 4 shows a simulated tumor with a final cell count of 10,000. The top-left, top-right, and bottom-left panels are projections along the 𝑧, 𝑥, and 𝑦, coordinates, respectively. The bottomright panel is an isometric view. No necrotic core was obtained for this small number of cells. Figure 4: Spheroid with a final cell number of 10,000. The top left, top right, and bottom left panels show projections along the 𝑧, 𝑥, and 𝑦 axes, respectively. The bottom right panel is the isometric view. Scale bars are 50 microns. No necrotic core was formed in a spheroid of this size based on the algorithm used to develop dead cells. Figure 5 and Figure 6 show the tumor geometries for 100,000 cells and 1,000,000 cells, respectively. The shape remains generally spherical, and a necrotic core forms. In each figure, Panel A shows the entire tumor, and Panel B shows the necrotic core only. 7 Figure 5: Spheroid with final cell number of 100,000. The panels correspond to the same projection directions as the corresponding panels in Figure 1. A is the whole spheroid, with 100 m scale bars; B is the necrotic core with 50 m scale bars. B Figure 6: Spheroid with final cell number 1,000,000. The panels correspond to the same projection directions as the corresponding panels in Figure 1. A is the whole spheroid, with 200 m scale bars; B is the necrotic core with 100 m scale bars. The desired cell counts used are 10,000, 100,000, 1,000,000, and 2,000,000. The spheroid containing 2,000,000 cells produced the most data and was used for further analysis. ANOVA testing of the data had a p-value of zero when tested with respect to doublings between trials, and the p-value of the data when tested with respect to trials between doublings was one. Alpha was 0.05. The average sphericity of the images was .94 with standard deviation of .01. Discussion The sizes of the simulated spheroids agreed with those simulated by Frieboes (2007) and Swanson (2003), but our simulations produced more spherical shapes. The spherical shapes agree with in vivo data by Sánchez (2001). 8 The program can be made more, accurate in terms of cell numbers, by factoring in the addition of nutrients This improvement will assist in developing reliable necrotic cores from the simulation. Diffusion equations can be examined to provide nutrient formation in the model as demonstrated in the literature (Mehta 2012, Venkatasubramanian 2006). Despite the simplicity of the algorithm, the simulation requires substantial processor time (e.g. 17 hours per simulation on an PC). However, the initial coding was not designed for speed, and small improvements can radically improve efficiency. Conclusion Simulated spheroids show strong relationships with tumors and diffusion model spheroids. The model is still relatively simple, but requires a large amount of processor time for a single simulation. Even with limitations, the method has potential to become more reliable with further research. References 1. Frieboes H., Lowengrub J., Wise, S. Zheng X., Macklin P., Bearer E., Cristinia V. 2007 Computer simulation of glioma growth and morphology, NeuroImage 37 S59–S70. 2. Mehta G., Hsiao A., Ingram M., Luker G., Takayama S., 2012. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. Journal of Controlled Release. 3. Sánchez C, de Ceballos M., del Pulgar T., Rueda D., Corbacho C., Velasco G., GalveRopherh I., Huffman J., Ramón S., Guzmán M., 2001. Inhibition of Glioma Growth in vivo by selective activation of the CB2 cannabinoid receptor. Cancer Research 61, 57845789. 4. Swanson K., Bridge C., Murray J.D., Alvord E., 2003. Virtual and real brain tumors: using modeling to quantify glioma growth and invasion. Journal of Neurological Sciences. 216, 1-10. 5. Venkatasubramanian R., Henson M., Forbes N., 2006. Incorporating energy metabolism into a growth model of multicellular tumor spheroids. Journal of Theoretical Biology. 242, 440-453. 9 Microfluidic Design and Platelet Adhesion on Charged Substrates Students: Josuha Kays and Max Henry Mentor: Steven A. Jones Introduction Thrombosis-related diseases such as Deep Vein Thrombosis (DVT) and Pulmonary Embolisms (PE) cause between 350,000-600,000 hospitalizations a year, including at least 100,000 deaths.1 The formation of a thrombus begins when the endothelium is damaged and blood is exposed to both collagen and tissue factor released from the deteriorating endothelium cells. This exposure activates platelets in the blood, and activation causes the platelets to stick and form a clot with fibrinogen fibers.2 Platelet activation occurs quickly through a positive feedback mechanism that causes other platelets to activate, while also being limited by platelet inhibitors such as Nitric Oxide (NO) or L-arginine.3 Together, the interactions between the positive feedback and negative feedback responses allow localized clotting to occur without coagulating all blood in the body. However, if a thrombus forms and breaks off from the endothelium into the blood stream, DVTs and PEs can occur and be life-threatening. Prevention of thrombosis depends on understanding blood coagulation mechanisms and platelet adhesion dynamics. Though research has been conducted on much of the above mechanisms in thrombus formation, there is still a great need for a physical model of platelet adhesion in human blood vessels. While much work has examined the relationship between a platelet’s local chemical environment and its activation, it is expected that platelet activation will depend also on the history of the platelet’s environment. In addition, it is expected that platelet activation and adhesion downstream of a region of platelet activation will be enhanced by the agents that activated platelets release. Experiments were therefore performed in which platelet-rich plasma flowed over an interface between two different surfaces, to determine whether the direction of flow, and hence a change in upstream/downstream surface configuration, altered the adhesion. Methods The microchannel design is shown in Figure 7. The LbL process is used to coat a glass slide with adhesion proteins. The slide is then covered with a rubber gasket that provides cutouts that act as flow channels. The inlets and outlets are provided through threaded holes that are drilled in the upper Plexiglas cover. Flow is generated with a syringe pump. The entire assembly was held together with a nylon spring clamp. 10 Figure 7: Microchannel design. A glass slide is covered with a silicone gasket into which channels are cut. A Plexiglas cover interfaces the two inlets and two outlets for flow. Dynamic LbL was conducted according to Lopez’s protocol4 for the first five polyelectrolyte bilayers of poly(diallyldimethylammonium chloride) (PDDA) and poly(styrene sulfonate) (PSS). This was followed by three bilayers of PDDA alternated with fibrinogen, where the solutiosn were passed through the half covered silicone mask at 120 ml/hr. Between layers, the channels were rinsed with phosphate-buffered saline for five minutes. Citrated whole bovine blood was spun at 1000 g for 60 minutes in a Hermle Labnet Z 323 centrifuge with Histopaque 1083 gradient buffer. This PRP was diluted to 50% in PBS solution (pH 7.5) and 1 ml of filtered acridine orange solution (1 mg/ml) was added, taking care to avoid light exposure. The full length channel mask was then placed on the glass slide, and the pre-stained PRP was pushed through at 20 ml/hr for 10 minutes, after which a 5% solution of glutaraldehyde in PBS was used to fix the cells for 10 min, though this was later reduced to 2.5% after knowledge of glutaraldehyde’s autofluorescence and ability to crosslink with fibrinogen was discovered. PBS was used to rinse for 5 minutes and the slides were allowed to air dry at room temperature. All slides were placed in aluminum foil-covered Petri dishes and refrigerated between 4-8° C when not in use. In this setup, 4 slides (8 channels) were run with the PRP contacting the fibrinogen side first, while another 4 slides had the PRP contacting the PDDA side first. Slides were imaged in both FITC and TRITC filtered light with a 10x objective Olympus microscope. Approximately 15 photos could be taken of each half of the channels, though care was taken to avoid false positive “drying rings” and the inlets and outlets of the channels. Images of the negative control – a layered slide before exposure to PRP – were also taken. The images were converted to gray scale then processed through a threshold to remove background fluorescence. These images were than analyzed with MATLAB to calculate percent surface coverage and each half channel was averaged together. Individual slide averages and the sum of slide averages were compared between fibrinogen and PDDA. Data collected for both directions of flow was corrected by removing the background fluorescence as determined by the closedshutter control shots. Data points above and below three standard deviations were removed. 11 Results Atomic force microscopy was used to examine the interface between the fibrinogen and PDDA coating. The interface is readily visible in Figure 8 as a sudden change in the surface height. Figure 8: AFM Hybrid d-LbL Fibrinogen/PDDA interface. Figure 9 shows the percentage of surface covered by platelets (represented by Tritc fluorescence), averaged over individual slides, for the case where flow crosses the fibrinogen half of the slide first and then the PDDA half. Figure 10 shows the percentage of surface cover for flow in the opposite direction. Figure 9: Platelet surface coverage on individual slides for flow passing from fibrinogen to PDDA 12 Figure 10: Platelet surface coverage on individual slides for flow passing from PDDA to fibrinogen. Figure 11 shows the average percent surface area coverage over all slides. Although a slight decrease in adhesion is seen on the PDDA surface, as viewed through the Tritc filter, the difference was not statistically significant. 2 Fibrinogen to PDDA Fibrinogen PDDA 3 Surface Area Coverage (%) Surface Area Coverage (%) 3 2 1 PDDA to Fibrinogen Fibrinogen PDDA 1 0 0 TRITC FITC TRITC FITC Figure 11: Surface coverage averaged over all slides. Left: fibrinogen to PDDA. Right: PDDA to fibrinogen. Discussion Although the mean value for surface coverage was smaller for PDDA than for fibrinogen, the difference was not statistically significant. The lack of significance was unexpected because fibrinogen is highly thrombogenic, yet PDDA’s thrombogenicity has not been noted in literature; Jirouskova et al. stated that polymer charge has no effect on adhesion and generally causes mild activation.5 However, this statement conflicts with previous work that indicated positively charged vessel walls induce coagulation, and negatively charged walls prevent coagulation.6. It is possible that other proteins in the PRP adsorbed to the PDDA surface, thus platelet adhesion 13 was not to PDDA but to an adsorbed surface on the PDDA. It is also possible that the Plexiglas surface contributed to platelet activation7. It may be useful to coat the Plexiglas in future studies. An F test demonstrated an increase variability between the forward and reverse flow slides. No previous research has yielded this result. Conclusion The AFM results demonstrated that the method for construction of the two-component channel surface was successful. No significant difference was found in platelet adhesion between the two surfaces. Improvements in the experimental setup include further refinements to reduce leakage, recalcification of the blood, and use of (negatively charged) PSS instead of (positively charged) PDDA. Finally, tests should be conducted to see what (if any) proteins are adsorbing to the biomaterial surfaces (PDDA or PSS). References 1. Galson, S.K. “The Surgeon General’s Call to Action to Prevent Deep Vein Thrombosis and Pulmonary Embolism,” http://www.surgeongeneral.gov/library/calls/deepvein/index.html 2. Z.M. Ruggeri, (1997) "Mechanisms initiating platelet thrombus formation." Thromb Haemost, vol. 78, no. 1, pp. 611-616, July 1997 3. Eshaq, R. (2006) “The Effect of the Local Concentrations of Nitric Oxide and ADP on Platelet Adhesion and Thrombus Formation.” 2006, Master of Science Engineering Thesis. Louisiana Tech University, Ruston, LA. 4. Lopez, J (2010) “An Improved Layer-By-Layer Self-Assembly Technique to Generate Biointerfaces for Platelet Adhesion Studies: Dynamic LbL.” Dissertation, Doctor of Philosophy, College of Engineering and Science, Louisiana Tech University. 5. Jirouskova, M., Bartunkova, J., and Smetana, K. (1997) "Comparative Study of Human Monocyte and Platelet Adhesion to Hydrogels in Vitro – Effect of Polymer Structure." Journal of Materials Science: Materials in Medicine. 6. Sawyer, P. and Srinivasan, S. (1972) “The role of electrochemical surface properties in thrombosis at vascular interfaces: cumulative experience of studies in animals and man.” Bull N Y Acad Med. 1972 February; 48(2): 235–256. (1997): SpringerLink. 7. Wang, Y. et al. "Effects of the Chemical Structure and the Surface Properties of Polymeric Biomaterials on Their Biocompatibility." SpringerLink. Springer Science Business Media, Aug. 2005. 14 The Effect of Growth Factors on Osteoblast Maturation in HNT-Loaded Alginate Hydrogels Student: Kanesha Hines Mentor: David K. Mills Introduction Hydrogels are hydrated polymer materials that form beads under defined conditions [1]. They can be synthetic or natural and are used to mimic the extracellular matrix in the body. They are made up of hydrophilic polymer chains that crosslink to form the beads. Hydrogels are biocompatible and easy to manufacture. Their composition is easily controlled and manipulated. Alginate hydrogel is often used because of its low toxicity and its ability to gel. Its medical applications include drug delivery, drug stabilization, and cell encapsulation. Hydrogels can be loaded with cells and implanted in wounds to decrease healing time. Halloysite nanotubes (HNTs) are used as nanocontainers for sustained drug release. HNTs are hollow tubes made from the naturally occurring aluminosilicate. They can be loaded with a diverse set of growth factors for sustained release [2]. In this study, Bone Morphogenic Proteins (BMPs) 4 and 6 were loaded into halloysite nanotubes. BMPs are proteins that stimulate ectopic bone growth and are members of the Transforming Growth Factor Beta (TGF-ß) superfamily. BMP-4 is a protein that stimulates ectodermal tissue differentiation. BMP 6 induces osteoblast differentiation in mesenchymal stem cells and subsequent osteogenesis. The effects of these agents on osteoblasts, cells responsible for bone tissue formation during fracture repair, are examined. Methods Preparation of the Alginate Beads All of the hydrogels were made with 2% w/v sodium alginate and 1% w/v HNTs loaded with BMP in saline. The HNTs were sterilized with 70% ethanol and vacuum loaded with BMP under sterile conditions. The BMP 4 solution was prepared with 100 mM acetic acid. 100 ml was removed and put into the BMP-4-labeled tubes and filled with 900 ml of water. 20 ml of the 1000 ml solution was then moved to another tube and 980 ml of water was added. For BMP-6, a 20 mM acetic acid solution was made. 20 µl was removed from the solution and put into the BMP 6 tube and then transferred to another tube and 980 µl of water was added. For the BMP-4 and 6 tubes, 0.05 g of BMP 4 and BMP 6 doped HNTs were used The final composition of each alginate bead was 0.2 g of alginate plus 0.1 g BMP doped HNTs in 10 ml of NaCl. The osteoblasts were then added to each BMP solution. A 1% w/v calcium chloride solution was made. 3- 12 well plates were labeled with the date of fixing and which BMP solution was present. The calcium chloride solution was added to every well and the BMP solutions were loaded into 27 g syringe and 6 drops were added to each well. The beads formed immediately upon contact with the calcium chloride. The beads were left to solidify for 15 minutes. The calcium chloride was then removed by pipette and replaced with DMEM medium. 15 The plates were then placed in the incubator and cultured for 28 days with samples fixed on days 7, 14, 21, and 28. Fixing the Samples To fix the samples, 3 freezing tubes, 3 large vials, and 9 (3 for each plate) medium vials were needed. Each tube was labeled with the assay and fixing date. The medium was removed from all the well of the labeled fixing date and 2 ml of medium was placed in each of the BMP-4, BMP-6, and BMP-4 and 6 tubes for freezing. Three beads from each BMP solution were placed in large tubes with sodium citrate to dissolve the beads. The rest of the beads were placed in the medium tubes based on BMP solution with ethanol. The beads were left in the ethanol for 10 minutes and then the ethanol was poured off. The freezing tubes were placed in the −80°C freezer and the medium and large vials were placed in the 4 °C refrigerator. This process was completed on day 7, 14, 21, and 28. Staining Fixed samples were stained for brightfield microscopy. The beads were placed in three, new 12well plates for the staining. Samples were stained with Von Kossa, Alcian Blue, and Picrosirus Red following standard staining procedures. After staining the beads were cut with a blade, they were mounted on microscopic slides. Mounting media and a cover slip was placed on each slide and each slide was observed and photographed using an Olympus BX51 microscope for analysis. Results Von Kossa staining demonstrates the degree of mineralization. For all three combinations, mineralization was low at Day 7 and increased at Days 14, 21, and 28, with nearly the entire field mineralized at Day 28. BMP 4 showed the most mineralization, followed by BMP 6. In the combination of BMP 4 and 6 there appeared to be a delay in the mineralization, but it followed the same pattern of increasing over time. Picrosirus red is an indicator of collagen. For each combination, the intensity of the stain increased initially, but then decreased. The peak occurred at Day 14 for BMP 4 and at Day 21 for BMP 6 (Figure 12) and the BMP 4/BMP 6 combination. Figure 12: Picosirius read staining for BMP 6. Left: Day 7. Right: Day 28. 16 Alcian blue stains acid glycosaminoglycans. At Day 7, staining was modest for BMP 4, stronger for BMP 6, and intense for the BMP 4/BMP 6 combination. In all cases, staining increased at Day 14. It peaked at Day 21 for BMP 4 and at Day 14 for BMP 6, the BMP 4/BMP 6 combination. Discussion The delay in mineralization for the combination of BMP 4/BMP 6 may occur because 4 and 6 provide different, potentially conflicting cell signals. The picrosirus red staining suggest that collagen increases initially but becomes mineralized at the later days, causing the decrease of the staining intensity. In BMP 4/BMP 6, delayed mineralization leads to a decrease of stain intensity. Thus, the decreased staining intensity for picosirius red mirrors the increased intensity for Von Kossa. The sulfated proteins that are present in collagen explain the decreases in alcian blue intensity in all 3 combinations. Conclusion All growth factor combinations enhanced osteoblast differentiation, functionality, and mineralization. BMP 4 led to the largest amount of bone mineralization. Future studies should use ELISA to identity and quantify bone protein. Additionally, we need to find an alternative method for the microscopy of hydrogels. The thickness of the gels degraded the microscopic images. References 1. Drury, Jeanie L. Mooney, David J. “Hydrogels for Tissue Engineering: Scaffold Design Variables and Applications” Biomaterials. Vol 24. 2003 Pg. 4337-4351. 2. Ruiling Qi , Rui Guo , Mingwu Shen , Xueyan Cao , Leqiang Zhang , Jiajia Xu , Jianyong Yu and Xiangyang Shi, “Electrospun poly(lactic-co-glycolic acid)/halloysite nanotube composite nanofibers for drug encapsulation and sustained release,” J. Mater. Chem., 20, 10622-10629, 2010. 17 Effects of Triton X-100, pH, and Ball Milling on Piezoelectric Nanocoating Student: Ivy Alexander Mentor: Chad O’Neal Introduction Lead zirconate titanate (PZT) is a piezoelectric material that is frequently used for sensors. The material produces an electric charge when it is stressed. Microcantilevers are frequently-used components of sensors, where the bending of the cantilever translates to an angular change, which can be detected optically. Alternatively, cantilevers can be coated with PZT in nanoparticle form to provide a charged-based signal. The coating process requires the nanoparticles to be dispersed in a solvent, and the quality of the coating depends on the content and properties of that solvent. Without dispersants, PZT nanoparticles agglomerate in the solution, leaving them clumped through Van der Waals forces and unevenly distributed . Dispersant induces electrostatic forces around the particles that counteract Van der Waals forces. Ball milling can produce smaller particles that improve the stability of the PZT particles in solution. Smaller particles reduce the effects of gravitational forces and Van der Waals forces while the improving electrostatic forces. This study examines the effect of dispersant concentration, pH, and ball milling on PZT nanocoatings. Methods The PZT powder was baked at 110 °C for 10 minutes to evaporate any water that could interfere with the dispersant’s effectiveness, combined with the X-100 dispersant, and then added to the solvent, a mock sol-gel solution of 4.405 mL 1-butanol and 0.595 mL propylene glycol . The final dispersant concentrations were 2, 4, 6, 8, and 10 mg/mL. The thickness of the coating layer, Hx, was then measured at 0.5, 1, 2, 4, 6, and 12 hours. After mixing, the colloidal mixture of power, dispersant, and solvent settled from a top layer within a cuvette, and the height of the lower layer (Hx) was measured as a function of time as a measure of the mixture’s stability. Results Effect of X-100 Surfactant Concentration Figure 13 shows the normalized layer thickness for three concentrations of X-100 dispersant. The thickness generally decreased (degraded) with time, and the most stable layer was obtained for the highest dispersant concentration. 18 1 Hx/H0 0.8 2.0 g/ml 1.0 g/ml 0.5 g/ml 0.6 0.4 0.2 0 0 1 2 3 4 5 6 Time (Hours) Figure 13: Normalized coating thickness as a function of time for three concentrations of X-100. Effect of pH The pH affects the electrostatic forces between particles. The pH was increased with 0.1 m/L NaOH or decreased with 1 m/L HCl to obtain a range from 3 to 11. The height ratios at 4 and 6 hours are presented as a function of pH in Figure 14. Higher and lower pH values tended to generate more stable layers, although the pH of 11 led to more degradation after 4 and 6 hours. 1 4 Hours 6 Hours Hx/H0 0.95 0.9 0.85 0.8 0 2 4 6 8 10 12 pH Figure 14: Height ratio as a function of pH. Balling/DLS/SEM The solvent, dispersant, and HCl were prepared in the ball milling canister, and the powders were then added. Particle size was measured with direct light scattering at 0. 2, 4, 6, 8, 12, 18, 19 24, and 48 hours. Samples were taken from the bottom, wall, and edge of the canister to examine the effect of canister position on particle size. SEM images were also taken. Various balling times points were taken some with and other without ultrasonication. Based on these readings, the average particle size, without any ball milling, fits into with the 200-800 nm range. Trial 1 the particle size at hours appears to be in accurate as the average is almost 800 nm while it is closer to 500 nm. This graph is based on all positionings (bottom, wall, edge) averaged to together to look at trend between the two trials. Due the first reading that appear to be almost opposite of each. Based on position trial one increased in particle size after 2 hours due to such a lower starting particle size. Then almost staying the same size after 4 hr (expect for the bottom). All decrease in particle size at 6 hr. Compared to trial 2 the opposite trends can be seen at hours and 6 hr. Ball milling did not consistently reduce particle size. It is suspected that the solution became unstable during ball milling resulting in particle agglomeration. This theory is back by a study done by Tralpho [1] which showed that excessive milling times led to aggregation of the particles. Our SEM images demonstrated this phenomenon (Figure 15). Figure 15: SEM images taken after 0 hours (left) and 6 hours (right) of ball milling. Aggregation is observed at 6 hours. Conclusions Triton X-100 improved the stability of PZT solutions. Acidic and basic solutions tended to be more stable. Ball milling did not consistently reduce the effective particle size because it tended to lead to aggregation. References [1] N. Traiphol, "Effects of ball milling time and dispersant concentration on properties of a lead," Journal of Ceramic Processing Research. vol. 8, No. 2, pp. 137-141, 2007. 20 21 of 25 Modified Silver Nanoparticles for Bacterial Infection Treatments Students: Megan Livingston and Christopher Ramirez Mentors: Dennis P. O’Neal and Sven Eklund Introduction Silver can act as an antimicrobial agent (Percival, 2005; Guzman, 2008). It has been infused into polymers and surfaces to prevent the bacterial adhesion and colonization of implantable medical devices (Furno, 2004). Silver nanoparticles (AgNPs) have been proposed as a direct treatment for infection (Jena, 2012). For this application, the nanoparticles must be sized such that they are cleared rapidly enough to prevent toxicity, yet slowly enough to provide time to affect pathogens. AgNPs must also be modified with compounds, such as sodium dodecyl sulfate (SDS) (Tillman, 2004), to prevent rapid degradation. This project investigated the manufacture, identification, modification, and stability of modified AgNPs and examine their potential oligodynamic effect on yeast cell cultures. Methods To generate the nanoparticles, 60 mL of 1 mM silver nitrate was mixed with 0.035g of SDS and then reduced with 180 mL of 2 mM sodium borohydride (>99% purity, Sigma Aldrich) under 1 minute of heavy stirring. The suspension was then allowed to sit overnight, and was then purified with a 10,000 mW dialysis cassette (Thermo Scientific). The DI water was replaced every 3 hours, and then allowed to dialyze overnight. AgNPs were concentrated via a rotory evaporator to bring the concentration to ~1 mM of Ag for 200. For the yeast cell cultures, the solution was only purified and not concentrated. To determine stability in the yeast cell media, a solution of 50% SDS AgNP and 50% media was made and a UV-Vis spectrophotometer (Evolution 60, Thermo Scientific) was used to analyze the nanoparticles. The growth media used was YPD (1% yeast extract, 2% glucose, 2% peptone) with a pH of ~6.5-7.0. To examine oligodynamics in yeast cell cultures, 100 µL saturated culture (Candida albicans strain 3153) was added to 50 ml YPD. When the optical density was ~0, ten vials were each filled with 4 mL of yeast cell culture solution. Five control vials were filled with a PBS solution containing the same %wt of SDS as the original solution (~1.5 mg for 10 mL of PBS) in amounts of 50, 100, 200, 500, and 1000 μL, respectively. The remaining five vials were filled with the purified AgNP solution with the same volumes. Vials were placed in a shaker (Max Q 4000, Barnstead) at 37 ⁰C for 2 hours. One mL of solution was then extracted from each of the ten vials to measure the optical density. Another set of extractions and measurements was also taken after 4 hours. Results SEM Images of SDS-AgNPs The SEM images in Figure 1 confirm the synthesis of AgNPs of somewhat uniform distribution of diameters between 5 and 15 nm (Figure 16). The small particle size is likely the result of 22 of 25 SDS acting as a physical barrier to agglomeration of the particles to larger sizes. It is proposed that NP size distribution might be controllable through changes in SDS concentration. Figure 16: Scanning electron micrographs of AgNPs on a silicon substrate. SDS-AgNP Stability in Media Over Time The UV-Vis spectral data in Figure 17 demonstrates the stability of AgNPs in media over the course of an hour. The height of the peak is directly related to the quantity of nanoparticles that absorb light at that wavelength. The height of the peak declines at a decreasing rate, indicating that in media, the SDS-AgNPs remain relatively stable for clinically relevant times. 1.4 Absorbance 1.2 0 min 30 min 60 min 1 0.8 0.6 0.4 0.2 0 0 200 400 600 800 Wavelength (nm) Figure 17: Spectra of SDS AgNPs in media showing the stability with time SDS-AgNP Stability in Phosphate Buffer Saline Solution The spectral data in Figure 18 show the effect of phosphate buffered saline (PBS) on the SDSAgNP solution. Absorbance peaks at higher wavelengths are indicative of larger nanoparticles, 23 of 25 while the height of the peak indicates quantity. At 0 hours, peaks exist for nanoparticles at 400 nm and 650 nm. The larger nanoparticles fall out of solution more rapidly, so the 650 nm peak disappears. However, the smaller, more stable nanoparticles persist for a longer period of time, even after exposure to a physiologic pH. 0.7 Absorbance 0.6 0 Hours 1 Hour 2 Hours 3 Hours 4 Hours 0.5 0.4 0.3 0.2 0.1 0 0 200 400 600 800 Wavelength (nm) Figure 18: UV-Vis spectra of Ag-SDS NPs in PBS over four hours Oligodynamics in Yeast Cell Cultures Figure 19 shows the optical density for Candida albicans strain 3153 after 4 hours, as a function of the added volume of AgNP solution. Higher optical densities indicate more bacteria. For the control cases, an equal amount of polymer solution was added. The purified SDS-AgNP solutions yielded lower optical densities than their control counterparts over four hours. This indicates that aside from the SDS modifier, the Ag present in the solution may have yielded an oligodynamic effect by lessening bacterial growth. 24 of 25 0.8 Control Growth Ave. Optical Density 0.7 AgNP Growth Ave. 0.6 0.5 0.4 0.3 0.2 0.1 0 1 2 3 4 5 Volume (L) Figure 19: Averages of SDS-AgNPs and SDS-Control Optical Densities for Total Growth Time Conclusion SDS-AgNPs were stable in the culture media and in PBS, and they yielded an oligodynamic effect at a physiologic pH in a protein-rich environment. However, the silver nanoparticles were smaller than desired, and the effect of SDS concentration on particle size should be further examined. The stability of the particles in blood must also be directly evaluated. Also, the toxicity of SDS on humans needs to be examined. Bibliography Furno, Franck, Kelly Morely, Ben Wong, Barry Sharp, et al. "Silver nanoparticles and polymeric medical devices: a new approach to prevention of infection?" Journal of Antimicrobial Chemotherapy, 54, 6. <http://jac.oxfordjournals.org/content/54/6/1019.short>. Guzman, Maribel, Jean Dille, and Stephan Godet. "Synthesis of Silver Nanoparticles by Chemical Reduction Method and Their Antibacterial Activity," World Academy of Science, Engineering and Technology, 43. Jena, Prajna, Mohanty Soumitra, et al. "Toxicity and antibacterial assessment of chitosan coated silver nanoparticles on human pathogens and macrophage cells," International Journal of Nanomedicine, 2012, 7 Percival, S.L., P.G. Bowler, and D. Russell. "Bacterial Resistance to Silver in Wound Care," Journal of Hospital Infection. Tillmann, Patricia. Stability of Silver Nanoparticles in Aqueous and Organic Media. Howard University, 2004. <http://www.nnin.org/doc/2004NNINreuTillmann.pdf>. 25 of 25