Section6.5_HW
advertisement

STAT 875 homework for Section 6.5 with partial answers
1) Problem #1 on p. 468 – Parts of this problem have been done in the class notes. Below is some
additional work for this problem.
For part c:
> mod.glmm.1s.i <- glmer(formula = good ~ distance + (1|kicker) +
(0+distance|kicker), nAGQ = 1, data = kick, family = binomial(link = "logit"))
> summary(mod.glmm.1s.i)
Generalized linear mixed model fit by maximum likelihood ['glmerMod']
Family: binomial ( logit )
Formula: good ~ distance + (1 | kicker) + (0 + distance | kicker)
Data: kick
AIC
783.0929
BIC
logLik
804.1406 -387.5465
deviance
775.0929
Random effects:
Groups
Name
Variance Std.Dev.
kicker
(Intercept) 0.07566 0.2751
kicker.1 distance
0.00000 0.0000
Number of obs: 1425, groups: kicker, 34
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 5.87481
0.33494
17.54
<2e-16 ***
distance
-0.11680
0.00846 -13.81
<2e-16 ***
--Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr)
distance -0.948
The estimated model is logit(ˆ ik ) 5.87 b0i (0.116 b1i )distanceik where b0i is a random
draw from a normal distribution with mean 0 and variance 0.076 and the b1i are all 0 (they are
random draws from a normal distribution with mean 0 and variance 0).
The LRT p-value using the chi-square distribution approximation is 0.5000. The bootstrap pvalue calculation is shown below.
> numb.sim <- 2000
> sim.H0 <- simulate(object = mod.glmm.1, nsim = numb.sim, seed = 8512)
> start.time <- proc.time()
> LRT.star <- numeric(length = numb.sim)
> for (i in 1:numb.sim){
mod.glmm.1 <- glmer(formula = sim.H0[[i]] ~ distance + (1|kicker), nAGQ = 1,
data = kick, family = binomial(link = "logit"))
mod.glmm.1s.i <- glmer(formula = sim.H0[[i]] ~ distance + (1|kicker) +
(0+distance|kicker), nAGQ = 1, data = kick, family = binomial(link =
"logit"))
mod.fit.wo <- glm(formula = sim.H0[[i]] ~ distance, data = kick, family =
binomial(link="logit"))
1
#print(i)
LRT.star[i] <- -2*(logLik(mod.glmm.1) - logLik(mod.glmm.1s.i))
}
> curve(expr = dchisq(x = x, df = 1), add = TRUE, col = "red")
> abline(v = LRstat.slope, col = "blue")
> summary(LRT.star)
Min.
1st Qu.
-0.000021 0.000000
Median
0.000000
Mean
0.210000
> mean(LRT.star >= LRstat.slope)
[1] 0.445
3rd Qu.
0.122800
Max.
4.791000
# p-value
> end.time <- proc.time()
> save.time <- end.time-start.time
> cat("\n Number of minutes running:", save.time[3]/60, "\n \n")
Number of minutes running: 57.08217
Notice the minimum -2log() value is a little less 0. I am not concerned about this because of
the numerical approximations used in evaluating the likelihood function and the minimum value
is still very close to 0. Also, it is interesting to note the highest bar in the histogram appears to
2
be left of 0. This is just likely due to R choosing the histogram classes so that first class
represents those values less than or equal to 0.
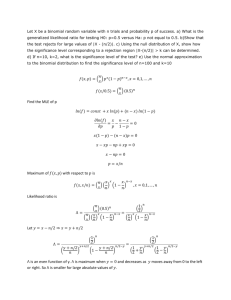
2) Consider the simple GLMM model logit(i ) 0 bi where Yi is a binomial random variable for the
ith item in the sample (ni = 10 trials) and bi ~ N(0, 2 ) for i = 1, …, n. The responses Yi are
independent for i = 1, …, n. The bi are independent as well. Complete the following:
a) Interpret what the model represents at the true probability of success and relate this to the
overdispersion discussion of Section 5.3.
b) Simulate n = 1000 observations from this model where 0 = 1 and = 2.
>
>
>
>
>
>
n <- 1000 # Number of binomial observations
beta0 <- 1
sigma <- 2
individual <- 1:n # Indicates observation number
numb.trials <- 10 # n_i
trials <- rep(x = numb.trials, times = n)
> # Simulate the response Y
> set.seed(8182)
> b <- rnorm(n = n, mean = 0, sd = sigma)
> pi.i <- plogis(beta0 + b)
> y <- rbinom(n = n, size = numb.trials, prob = pi.i)
> head(y)
[1] 10 0 3 1 10 10
c) Estimate the corresponding model to the simulated data. Compare the estimated values of 0
and to their true values. Use 20 quadrature points.
> # Estimate model with 20-point quadrature
> mod.fit20 <- glmer(formula = y/trials ~ 1 + (1|individual), nAGQ = 20, weights =
trials, family = binomial(link = "logit"))
> summary(mod.fit20)
Generalized linear mixed model fit by maximum likelihood ['glmerMod']
Family: binomial ( logit )
Formula: y/trials ~ 1 + (1 | individual)
AIC
2795.171
BIC
logLik
2804.987 -1395.586
deviance
2791.171
Random effects:
Groups
Name
Variance Std.Dev.
individual (Intercept) 3.966
1.992
Number of obs: 1000, groups: individual, 1000
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.98927
0.06966
14.2
<2e-16 ***
--Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
The estimated model is logit(ˆ i ) 0.9893 bi where bi ~ N(0,1.9922). As would be expected
with a large sample size, these estimates are fairly close to their true values.
3
d) Examine what happens to the estimated model with more and less points of quadrature.
e) The purpose of this part is to examine how similar models can be estimated using pre-Section
6.5 methods.
i) Estimate logit(i ) 0 using maximum likelihood like in Chapter 2. Is there evidence of
overdispersion?
Yes! For example, see the deviance/df statistic.
ii) Estimate logit(i ) 0 using a quasi-binomial model. Similar to the quasi-Poisson
regression model, a parameter allows for the variance to be nii(1 – i) rather than nii(1
– i) only.
iii) Compare the models in i) and ii) to the model estimated in part c).
4