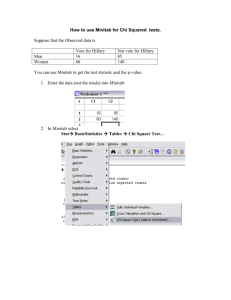
Chi square and Fisher exact test
advertisement

Categorical data Previously, we've analyzed data using t-tests, anova, or linear regression for which the dependent (response, y) variable was quantitative. These methods are usually not appropriate when the dependent variable is categorical: live, die cured, not cured success, failure disease, no disease, republican, democrat, other none, mild, moderate, severe Sometimes we have only two levels of the categorical response (success, failure) and sometimes we have more than two levels (republican, democrat, other). Sometimes there is no order to the levels, and sometimes there is an order (none, mild, moderate, severe). We use categorical data analysis methods to analyze categorical response variables. Among the commonly-used categorical methods are the following. Frequency tables Chi-square test Fisher exact test Cochran-Mantel-Haenszel (CMH) test Logistic regression We'll cover frequency tables, chi-square test and Fisher Exact Test in some detail. We won't cover CMH and logistic regression, but you should be aware they exist if you do analysis of categorical data. Frequency and contingency tables Frequency tables or contingency tables provide counts of the number of individuals who experience some event (such as cancer) conditioned on some property of the individual (such as having a specific gene mutation or not). mutation.data = matrix(c(5,35,20,580), nrow=2, byrow=T) colnames(mutation.data) = c("Cancer", "No cancer") rownames(mutation.data) = c("Mutation ", "No mutation") mutation.data Mutation No mutation Cancer No cancer 5 35 20 580 Tables that show frequencies or counts have several names. Frequency table: shows the frequency (and sometimes percent) of each category. 2*2 table indicates a table with 2 rows and 2 columns, as in the example above. r*c indicates a table with r rows and c columns. These tables are also sometimes called contingency tables. The word contingency just indicates that the counts are contingent on (dependent on) some property of the individual, such as cancer/not versus mutation/not. Chi-square and Fisher Exact tests Suppose we wish to know if a drug is effective at curing a disease. We perform an experiment, and observe the following result. Observed outcome: Drug Placebo Cured 50 0 Not cured 0 50 In this case, we observe that all the patients who received the drug were cured, while none of the patients who received the placebo were cured, The null hypothesis is that the drug doesn't have any effect, that is, there is no association between treatment and cure. Ho: There is no association between treatment and cure. If there is no association between drug and cure, we expect half of each group to be cured, and half not to be cured. Expected result if the null hypothesis is true: Drug Placebo Cured 25 25 Not cured 25 25 We use the chi-square test (or the Fisher Exact test) to test for an association between treatment and cure. The chi squared test examines the difference between the observed outcome and the outcome expected by chance, and tells us the probability that the observed outcome would have occurred in the absence of any true association (between drug and cure in this case). cure.data = matrix(c(50,0,0,50), nrow=2, byrow=T) colnames(cure.data) = c("Cured", "Not cured") rownames(cure.data) = c("Drug", "Placebo") cure.data Drug Placebo Cured Not cured 50 0 0 50 We use the R function chisq.test() to test for an association between treatment (drug vs placebo) and outcome (cure, not): chisq.test(cure.data) Pearson's Chi-squared test with Yates' continuity correction data: cure.data X-squared = 96.04, df = 1, p-value < 2.2e-16 The p-value is p < 2.23-16, which is essentially zero. The p-value is significant at alpha = 0.05, so we reject the null hypothesis that there is no association between drug and cure. Optional material: Calculation of the chi-square statistic and p-value In this section, we'll examine how the chi-square statistic and it's p-value are calculated. You don't have to know this to use the chi-square test, but it is helpful to understand the method, and also to understand when it can fail. You may recall the T statistics used in the t-test. T= Mean( placebo ) Mean(drug ) SEM ( placebo ) 2 SEM (drug ) 2 The T statistic is a function of the difference between the mean of the drug and the mean of the placebo. Notice that, if the drug has no effect, the expected value of the mean for drug group is just the mean of the placebo group. If the drug has no effect: mean of the drug (observed result) = the mean of the placebo (the expected result) If the drug has no effect: observed – expected = 0 The chi-square test follows the same logic. It compares the observed result (what we actually see) to the expected result if the null hypothesis is true (what we would see if there is no association). The chi-square statistic is a function of the differences between observed (O) and expected (E) number of events in each cell of the table. Suppose we have a table with g=2*2=4 cells. Observed outcome: Cured 50 0 Drug Placebo Not cured 0 50 Expected result if the null hypothesis is true: Cured 25 25 Drug Placebo Not cured 25 25 Here is the formula to calculate the chi-square statistic: (Oi Ei )2 Ei i 1 g 2 In our example, we have g=2*2=4 cells. chi-square = (50-25)2/25 + (0-25)2/25 + (0-25)2/25 + (50-25)2/25 =252/25 + 252/25 + 252/25 + 252/25 = 100 The p-value for the chi-square test is the probability that a chi-square value of 100 or greater would occur when the null hypothesis is true. The actual value of the chi-square statistic calculated by the R software is slightly smaller than 100, because it applies a correction factor called the "Yates continuity correction". The Yates continuity correction makes the estimated p-value more accurate. Optional material: How to calculate the expected value in each cell and get the pvalue using chi-square. The expected value for each cell is Eij = rowi total * columnj total / grand total, as follows. 1. Calculate the row totals, column totals, and grand total for the observed data Drug Placebo Column Total Cured 50 0 Not cured 0 50 50 50 Row Total 50 50 100 <= Grand total 2. Calculate expected value for each cell as (row total * column total / grand total) Drug Placebo Column Total Cured 25 25 Not cured 25 25 50 50 Row Total 50 50 100 <= Grand total The R function automatically calculates the expected count in each cell, so you don't need to. Chi-square test example Example: is there an association between cancer and smoking? Suppose we have data on 60 individuals who smoke and 60 who don't. We determine the number of each that have cancer. Smoke No smoking Cancer No cancer 40 20 20 40 What is the probability that we see the distribution if there is no association between smoking and cancer? smoking.data = matrix(c(40,20,20,40), nrow=2, byrow=T) colnames(smoking.data) = c("Cancer", "No cancer") rownames(smoking.data) = c("Smoke", "No smoking") smoking.data Smoke No smoking > Cancer No cancer 40 20 20 40 chisq.test(smoking.data) Pearson's Chi-squared test with Yates' continuity correction data: smoking.data X-squared = 12.0333, df = 1, p-value = 0.0005226 The p-value = 0.0005226 is significant at alpha = 0.05, so we reject the null hypothesis that there is no association between cancer and smoking. It is not required that the number of individuals in each group be the same. Example: is there an association between a mutation and smoking? Collect data on 40 individuals who have the mutation and 600 individuals who don't. Determine the number of each that have cancer. mutation.data = matrix(c(5,35,20,580), nrow=2, byrow=T) colnames(mutation.data) = c("Cancer", "No cancer") rownames(mutation.data) = c("Mutation ", "No mutation") mutation.data Mutation No mutation Cancer No cancer 5 35 20 580 > > chisq.test(mutation.data) Pearson's Chi-squared test with Yates' continuity correction data: mutation.data X-squared = 6.1301, df = 1, p-value = 0.01329 Warning message: In chisq.test(mutation.data) : Chi-squared approximation may be incorrect > The p-value = 0.01329 is significant at alpha = 0.05, so we reject the null hypothesis that there is no association between the mutation and smoking. Notice that we get a warning message: Warning message: In chisq.test(mutation.data) : Chi-squared approximation may be incorrect We get the error message because the chi-square test is an approximation to the Fisher exact test, and requires that certain assumptions are met. If the assumptions are not met, the chi-square p-value may be quite wrong. Usually, the assumptions are met if the expected number of observations in each cell of the table is greater than 5. 1. Calculate the row totals, column totals, and grand total for the observed data Cured Not cured Row Total Drug 5 35 40 Placebo 20 580 600 Column Total 25 615 640 <= Grand total 2. Calculate expected value for each cell as (row total * column total / grand total) Cured Not cured Row Total Drug 1.5625 38.4375 40 Placebo 23.4375 576.5625 600 Column Total 25 615 640 <= Grand total In this example, the expected number in the cell Drug/Cured is 1.56, so the chi-square approximation may not give us a reliable p-value. In these situations, we use the Fisher Exact test. Fisher Exact test The chi-square test is an approximate test that is fast and easy to calculate. It is based on the normal approximation to the binomial distribution. The approximation works reasonably well for large sample size, but the p-values are inaccurate for small sample sizes. By default, for 2x2 tables, R uses a version of the chi-square test with the Yates continuity correction that gives relatively accurate p-values even for small sample size. When the expected number of observations in any cell is small (less than 5), the Fisher Exact test is preferred. fisher.test(mutation.data) Fisher's Exact Test for Count Data data: mutation.data p-value = 0.01552 alternative hypothesis: true odds ratio is not equal to 1 95 percent confidence interval: 1.142297 12.258694 sample estimates: odds ratio 4.126956 In this example, the chi-square approximation with Yates continuity correction gave p=0.01329, while the Fisher exact test gives p=0.01552, so the results are similar. Here's another example where the results are different. fisher.data = matrix(c(2, 15,2,160), nrow=2, byrow=T) colnames(fisher.data) = c("Cancer", "No cancer") rownames(fisher.data) = c("Mutation ", "No mutation") fisher.data Mutation No mutation > Cancer No cancer 2 15 2 160 > fisher.test(fisher.data) Fisher's Exact Test for Count Data data: fisher.data p-value = 0.04561 alternative hypothesis: true odds ratio is not equal to 1 95 percent confidence interval: 0.7058128 152.8072330 sample estimates: odds ratio 10.37043 > chisq.test(fisher.data) Pearson's Chi-squared test with Yates' continuity correction data: fisher.data X-squared = 3.7327, df = 1, p-value = 0.05336 Warning message: In chisq.test(fisher.data) : Chi-squared approximation may be incorrect > In this example, the chi-square approximation give p=0.053, while the Fisher exact test gives p=0.0456. Optional material: Chi-square and Fisher tests for tables with more than two rows or columns The chi-square and Fisher exact tests are not limited to tables with only two rows or columns drug2.data = matrix(c(12, 33, 14, 51, 3, 39), nrow=3, byrow=T) colnames(drug2.data) = c("Relapse", "No relapse") rownames(drug2.data) = c("Placebo", "Dose A", "Dose B") drug2.data > drug2.data Relapse No relapse Placebo 12 33 Dose A 14 51 Dose B 3 39 > fisher.test(drug2.data) Fisher's Exact Test for Count Data data: drug2.data p-value = 0.04273 alternative hypothesis: two.sided > chisq.test(drug2.data) Pearson's Chi-squared test data: drug2.data X-squared = 5.8086, df = 2, p-value = 0.05479 In this case, the Fisher test is significant (p=0.04273) while the chi-square test is not (p=0.05479) The chi-square test was developed before we had fast computers. For most biomedical data sets, it is possible to run the Fisher Exact test in a reasonable amount of time. Examples using data frames For these examples we'll use the stroke data set from the ISwR package. install.packages("ISwR") library(ISwR) head(stroke) sex died 1 Male 1991-01-07 2 Male <NA> 3 Male 1991-06-02 4 Female 1991-01-13 5 Female <NA> 6 Male 1991-01-13 dstr age dgn coma diab minf han dead obsmonths 1991-01-02 76 INF No No Yes No TRUE 0.16339869 1991-01-03 58 INF No No No No FALSE 59.60784314 1991-01-08 74 INF No No Yes Yes TRUE 4.73856209 1991-01-11 77 ICH No Yes No Yes TRUE 0.06535948 1991-01-13 76 INF No Yes No Yes FALSE 59.28104575 1991-01-13 48 ICH Yes No No Yes TRUE 0.10000000 ?stroke A data frame with 829 observations on the following 10 variables. sex a factor with levels Female and Male. died a Date, date of death. dstr a Date, date of stroke. age a numeric vector, age at stroke. dgn a factor, diagnosis, with levels ICH (intracranial haemorrhage), ID (unidentified). INF (infarction, ischaemic), SAH (subarchnoid haemorrhage). coma a factor with levels No and Yes, indicating whether patient was in coma after the stroke. diab a factor with levels No and Yes, history of diabetes. minf a factor with levels No and Yes, history of myocardial infarction. han a factor with levels No and Yes, history of hypertension. obsmonths a numeric vector, observation times in months (set to 0.1 for patients dying on the same day as the stroke). dead a logical vector, whether patient died during the study. Example. Is there an association between sex and diabetes in this data set? mytable= xtabs(~ sex + diab, data=stroke) mytable chisq.test(mytable) fisher.test(mytable) diab sex No Yes Female 434 69 Male 288 28 > chisq.test(mytable) Pearson's Chi-squared test with Yates' continuity correction data: mytable X-squared = 3.932, df = 1, p-value = 0.04738 > fisher.test(mytable) Fisher's Exact Test for Count Data data: mytable p-value = 0.04509 alternative hypothesis: true odds ratio is not equal to 1 95 percent confidence interval: 0.3699738 0.9894478 sample estimates: odds ratio 0.6118643 Example. Is there an association between sex and death in this data set? mytable= xtabs(~ sex + dead, data=stroke) mytable prop.table(mytable,1) chisq.test(mytable) fisher.test(mytable) > mytable dead sex FALSE TRUE Female 189 321 Male 155 164 > prop.table(mytable,1) dead sex FALSE TRUE Female 0.3705882 0.6294118 Male 0.4858934 0.5141066 In these data, 62.9% of women and 51.4% of men die. Is this statistically significant? > chisq.test(mytable) Pearson's Chi-squared test with Yates' continuity correction data: mytable X-squared = 10.2779, df = 1, p-value = 0.001346 > fisher.test(mytable) Fisher's Exact Test for Count Data data: mytable p-value = 0.001122 alternative hypothesis: true odds ratio is not equal to 1 95 percent confidence interval: 0.4644489 0.8359763 sample estimates: odds ratio 0.6233559