Genomic Prediction for Rust Resistance in Diverse Wheat
advertisement

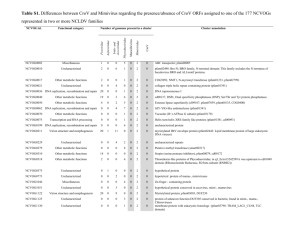
Genomic Prediction for Rust Resistance in Diverse Wheat Landraces Hans D. Daetwyler*, Urmil K. Bansal, Harbans S. Bariana, Matthew J. Hayden and Ben J. Hayes Theoretical and Applied Genetics, 2014 *Corresponding author: hans.daetwyler@depi.vic.gov.au Supplementary Information Table S1. List of Watkins genotypes (Triticum aestivum) used in study along with their country of origin Aus No. Country Aus No. Country Aus26410 Greece Aus27983 Soviet union republic Aus26414 Greece Aus27307 Soviet union republic Aus26429 Greece Aus27314 Australia Aus26431 Greece Aus27315 Australia Aus26438 Cyprus Aus27317 Australia Aus26491 Greece Aus27318 Australia Aus26526 Morocco Aus27320 Australia Aus26552 Portugal Aus27339 Australia Aus26558 Iran Aus27346 Yugoslavia Aus26559 Iran Aus27348 Yugoslavia Aus26580 Portugal Aus27354 Myanmar Aus26624 Spain Aus27355 Myanmar Aus26667 Tunisia Aus27356 Morocco Aus26673 Tunisia Aus27357 Morocco Aus27215 Afghanistan Aus27361 Spain Aus27225 Yugoslavia Aus27362 Spain Aus27905 Syria Aus27363 Spain Aus27229 Spain Aus27367 Soviet union republic Aus27230 Spain Aus27374 India Aus27247 Yugoslavia Aus27375 China Aus27251 Yugoslavia Aus27376 China Aus27252 Yugoslavia Aus27379 China Aus27255 Greece Aus27380 China Aus27270 Unknown Aus27382 China Aus27272 Unknown Aus27383 China Aus27279 Afghanistan Aus27389 China Aus27280 Afghanistan Aus27401 China Aus27284 Afghanistan Aus27402 China Aus27286 Afghanistan AUS27404 China Aus27294 Algeria Aus27405 China Aus27299 Algeria Aus27406 China 1 Aus27407 China Aus27843 India Aus27408 China Aus27848 India Aus27415 China Aus27851 India Aus27419 China Aus27852 India Aus27425 China Aus27853 India Aus27429 China Aus27854 India Aus27430 China Aus27856 India Aus27431 China Aus27857 India Aus27432 China Aus27858 India Aus27441 China Aus27861 India Aus27444 China Aus27869 India Aus27447 China Aus27873 Soviet union republic Aus27448 China Aus27878 Italy Aus27450 China Aus27881 Italy Aus27451 China Aus27882 Italy AUS27455 China Aus27885 Italy Aus27471 Yugoslavia Aus27894 Spain Aus27473 Yugoslavia Aus27899 Morocco AUS27474 Yugoslavia Aus27900 Morocco Aus27476 Yugoslavia Aus27902 Morocco Aus27477 Yugoslavia Aus27904 Spain Aus27492 France Aus27911 Spain Aus27506 France Aus27915 Spain Aus27524 Hungary Aus27916 Spain Aus27525 Hungary Aus27919 Spain Aus27800 Unknown Aus27921 Iran Aus27801 India Aus27928 Iran Aus27806 India Aus27930 Iran Aus27811 India Aus27931 Iran Aus27812 India Aus27940 Poland Aus27817 India Aus27949 Poland Aus27822 India Aus27950 Poland Aus27823 India Aus27953 Poland Aus27825 India Aus27955 Portugal Aus27826 India Aus27959 Portugal Aus27830 India Aus27960 Portugal Aus27832 India Aus27961 Portugal Aus27833 India Aus27962 Portugal Aus27834 India Aus27969 Portugal Aus27835 India Aus27970 Portugal Aus27836 India Aus27993 Yugoslavia Aus27838 India Aus27999 Italy Aus27841 India Aus28000 Spain Aus27842 India Aus28004 Soviet union republic 2 Aus28009 Egypt Aus28164 Iran Aus28011 Tunisia Aus28166 Iran Aus28012 Soviet union republic Aus28168 Iran Aus28014 Soviet union republic Aus28170 Iran Aus28015 Soviet union republic Aus28176 Iran Aus28020 Soviet union republic Aus28177 Iran Aus28022 Soviet union republic Aus28178 Iran Aus28028 Spain Aus28179 Iran Aus28042 Spain Aus28183 Iran Aus28044 Spain Aus28184 Iran Aus28050 India Aus28186 Iran Aus28052 India Aus28187 Iran Aus28055 India Aus28193 Portugal Aus28056 India Aus28194 Portugal Aus28058 India Aus28201 Portugal Aus28062 India Aus28202 Portugal Aus28066 India Aus28203 Portugal Aus28077 India Aus28205 Portugal Aus28081 India Aus28216 Yugoslavia Aus28082 India Aus28217 Yugoslavia Aus28083 India Aus28223 Spain Aus28086 India Aus28225 Spain Aus28089 India Aus28230 Turkey Aus28091 India Aus28232 Turkey Aus28095 India Aus28236 Tunisia Aus28096 India Aus28244 Bulgaria Aus28097 India Aus28245 Bulgaria Aus28099 India Aus28246 Bulgaria Aus28112 Iraq Aus28253 Greece Aus28115 Morocco Aus28254 Greece Aus28119 Morocco Aus28255 Yugoslavia Aus28124 Morocco Aus28261 Greece Aus28125 Morocco Aus28262 Yugoslavia Aus28126 Morocco Aus28264 China Aus28127 Morocco Aus28266 Ethiopia Aus28128 Morocco Aus28272 Soviet union republic Aus28138 Spain Aus28768 India Aus28139 Spain Aus28824 Yugoslavia Aus28141 Spain Aus29883 United Kingdom Aus28142 Spain Aus28266 Ethiopia Aus28146 Spain Aus28155 Spain Aus28160 Iran Aus28163 Iran 3 Table S2. The 40 SNPs with the largest absolute allele substitution effects for the second allele for leaf, stem and stripe rust ordered by genomic region. SNP effects presented as absolute values* SNP name/rust SNPEffect Chromo some Map order Gene QTL wsnp_Ex_c7252_12452995 0.00281 1A 21.0 Uncharacterized wsnp_Ex_c42282_48901252 0.00259 1A 87.2 Unpublished wsnp_Ex_c12336_19687074 0.00244 1A 87.2 wsnp_Ku_c21364_31103315 0.00227 1A 102.3 wsnp_JD_c13384_13393159 0.00233 1A 113.8 wsnp_RFL_Contig3542_3718200 0.00293 1A 117.5 wsnp_BE444305A_Td_2_1 0.00290 1A 117.5 wsnp_Ex_c10631_17340809 0.00254 1A 155.3 Uncharacterized wsnp_BE637971B_Ta_1_20 0.00260 1B 9.6 Uncharacterized wsnp_Ex_rep_c102909_87976721 0.00269 1B 64.1 Uncharacterized wsnp_BF485168B_Ta_2_1 0.00230 1B 66.5 wsnp_Ex_c27176_36393952 0.00245 1B 101.9 wsnp_CAP12_c1337_682282 0.00249 1B 121.9 wsnp_Ra_c21104_30458226 0.00239 2A 4.1 wsnp_Ex_rep_c102538_87682273 0.00278 2A 134.6 wsnp_Ra_c8430_14289874 0.00257 2A 150.5 wsnp_Ex_c4272_7708423 0.00229 2B 44.0 Uncharacterized wsnp_Ra_c31353_40494768 0.00228 2B 188.8 Yr43, Yr44, Yr53 QTL56 wsnp_Ku_c15498_24122936 0.00288 2B 208.5 Uncharacterized QTL7 wsnp_Ex_c24135_33382700 0.00295 2B 208.7 wsnp_BE488779D_Ta_1_2 0.00248 2D 81.6 Uncharacterized wsnp_Ra_c16278_24893033 0.00236 3A 42.8 Uncharacterized wsnp_Ku_c40218_48484410 0.00258 3A 43.3 wsnp_Ex_c16615_25147492 0.00254 3A 69.2 Uncharacterized wsnp_Ex_c15269_23492289 0.00238 3A 83.2 Uncharacterized wsnp_BE446462D_Ta_2_1 0.00321 3B 0.7 wsnp_CAP11_c1214_700133 0.00263 3B 52.8 wsnp_JD_c9805_10591233 0.00268 3B 62.4 wsnp_Ex_c5074_9009245 0.00234 3B 88.1 wsnp_JD_rep_c49010_33257826 0.00259 3B 96.2 wsnp_BE490599A_Ta_2_1 0.00241 4A 67.0 wsnp_RFL_Contig3024_2910610 0.00243 4A 67.0 wsnp_BE443291A_Ta_2_1 0.00244 4A 67.0 wsnp_Ex_c4331_7808746 0.00237 4A 165.4 wsnp_RFL_Contig4475_5295587 0.00278 4A 178.6 wsnp_BE442666B_Ta_2_2 0.00291 4B 36.5 Uncharacterized wsnp_Ex_c14812_22928900 0.00240 5A 45.0 Uncharacterized wsnp_Ex_c12678_20148981 0.00251 5A 91.6 Uncharacterized Stripe rust QTL1 Uncharacterized Uncharacterized QTL2,3 Yr41 QTL2,3,4 Unpublished Yr30 QTL8 Uncharacterized Uncharacterized Yr51 QTL9 QTL2 4 wsnp_Ku_c12211_19780409 0.00242 5A 95.6 wsnp_Ex_c898_1739090 0.00238 5A 118.2 wsnp_Ex_c898_1738424 0.00237 5A 118.2 wsnp_BE499835B_Ta_2_5 0.00211 5B 23.3 wsnp_Ku_c21465_31217980 0.00246 5B 201.8 wsnp_Ku_c3151_5892200 0.00246 5B 205.0 wsnp_Ex_c1857_3498746 0.00230 5B 214.2 wsnp_Ex_rep_c68491_67318138 0.00236 5D1 6.0 wsnp_Ex_c3422_6283568 0.00234 5D2 34.6 wsnp_RFL_Contig2424_1966870 0.00236 6A 0.9 Uncharacterized wsnp_JD_c3441_4455541 0.00266 6A 45.3 Uncharacterized wsnp_CAP11_c1114_653767 0.00325 6A 45.7 wsnp_Ex_c2236_4189774 0.00290 6A 180.2 wsnp_Ex_c34597_42879693 0.00282 6A 180.2 wsnp_Ex_c7191_12352173 0.00255 6B 7.1 wsnp_Ex_c45081_50974769 0.00250 6B 48.8 wsnp_Ex_c25505_34771897 0.00289 6B 52.9 wsnp_Ex_c38198_45786860 0.00230 6B 59.5 wsnp_Ex_c2849_5262624 0.00228 6B 60.1 wsnp_Ku_c5623_9966516 0.00234 6B 95.0 wsnp_Ra_c35443_43984445 0.00231 6B 95.0 wsnp_CAP11_c1432_806102 0.00283 6B 105.2 wsnp_RFL_Contig308_2996797 0.00254 7A 5.6 Uncharacterized wsnp_Ku_c139_279238 0.00245 7A 40.6 Uncharacterized wsnp_Ex_c25644_34905425 0.00255 7A 51.2 wsnp_Ex_c11636_18742884 0.00243 7A 82.3 wsnp_Ex_c18352_27178687 0.00234 7A 82.3 wsnp_Ex_c15988_24402251 0.00235 7A 82.3 wsnp_Ex_c42836_49314564 0.00239 7A 82.3 wsnp_Ra_rep_c104968_88985755 0.00238 7A 87.2 wsnp_JD_c15333_14824351 0.00236 7A 92.8 wsnp_BQ160404A_Ta_1_1 0.00251 7A 116.5 Uncharacterized wsnp_Ku_c8497_14429303 0.00280 7B 31.6 Uncharacterized wsnp_CAP8_c3593_1773371 0.00236 7B 45.3 wsnp_Ku_c19037_28455905 0.00261 7B 57.4 wsnp_RFL_Contig1205_284961 0.00239 7B 57.4 wsnp_BG262287B_Td_2_5 0.00247 7B 108.7 wsnp_Ex_c38203_45790396 0.00175 1A 22.5 Uncharacterized wsnp_Ex_c6452_11213329 0.00150 1A 97.5 Uncharacterized wsnp_Ex_c20489_29564938 0.00143 1A 97.5 wsnp_BG606986A_Ta_2_1 0.00148 1A 114.3 wsnp_BG606986A_Ta_2_4 0.00140 1A 114.6 wsnp_JD_c20553_18260731 0.00133 1A 128.9 wsnp_Ku_c13768_21859275 0.00164 1A 153.5 wsnp_Ex_c9600_15889471 0.00163 1A 179.2 Yr47 Uncharacterized QTL3,5,7,10,11 Uncharacterized QTL12,13 Uncharacterized Uncharacterized QTL1,7,14,15 Uncharacterized QTL14 Uncharacterized QTL7 Yr39, Yr52 Leaf rust Uncharacterized 5 wsnp_Ex_c33654_42106735 0.00130 1B 22.9 wsnp_Ex_c14832_22953906 0.00130 1B 22.9 Lr44, Lr71 wsnp_Ex_c6022_10553838 0.00143 1B 38.2 wsnp_Ex_c4436_7981188 0.00143 1B 123.4 wsnp_Ex_c866_1684236 0.00167 2A 63.7 wsnp_Ra_c4503_8155485 0.00131 2A 150.5 Uncharacterized wsnp_Ku_c4042_7375053 0.00129 2B 80.8 Lr13, Lr23, Lr48 wsnp_RFL_Contig2666_2354537 0.00127 2B 110.8 wsnp_CAP8_c7453_3441062 0.00139 2B 149.4 wsnp_BF291674B_Ta_2_1 0.00140 2B 149.4 wsnp_Ku_c12037_19549078 0.00143 2B 165.9 wsnp_Ex_c16425_24923685 0.00131 2B 185.7 wsnp_JD_c4343_5462565 0.00133 2B 191.0 wsnp_Ex_rep_c103064_88104690 0.00140 2B 230.1 wsnp_Ex_c25311_34578436 0.00146 2D 113.1 Uncharacterized wsnp_Ra_c16278_24893033 0.00175 3A 42.8 Uncharacterized wsnp_Ex_c24432_33676448 0.00158 3A 42.8 wsnp_Ku_c40218_48484410 0.00170 3A 43.3 wsnp_Ku_c3286_6111264 0.00151 3A 102.9 wsnp_Ex_c15100_23284292 0.00153 3A 102.9 wsnp_Ex_c9510_15761235 0.00153 3A 102.9 wsnp_Ex_rep_c104125_88923836 0.00169 3A 162.4 wsnp_Ex_c6223_10857649 0.00133 3B 80.9 wsnp_Ex_c18624_27492167 0.00139 3B 80.9 wsnp_Ex_c2352_4405961 0.00167 4A 136.3 Uncharacterized wsnp_JD_c17976_16616890 0.00158 4B 123.4 Uncharacterized wsnp_JD_rep_c51623_35119179 0.00153 4D 20.6 Uncharacterized wsnp_Ex_rep_c67296_65839761 0.00165 4D 22.4 wsnp_Ex_c34252_42593715 0.00142 4D 69.9 Lr67 wsnp_Ku_c9559_16000086 0.00133 5A 17.4 Uncharacterized wsnp_Ex_c49211_53875600 0.00132 5A 83.0 wsnp_Ex_c49211_53875575 0.00132 5A 83.0 wsnp_Ex_rep_c101994_87256479 0.00149 5A 136.7 wsnp_Ex_c23787_33024604 0.00146 5A 140.6 wsnp_BF201102B_Ta_2_5 0.00131 5B 23.6 Lr52 wsnp_Ex_c214_422365 0.00142 5B 39.4 Uncharacterized wsnp_Ex_c24577_33826666 0.00147 5B 57.1 wsnp_BE497820B_Ta_2_3 0.00135 5B 79.1 wsnp_BE495277B_Ta_2_5 0.00142 5B 101.6 wsnp_JD_rep_c63083_40243538 0.00137 5B 127.0 wsnp_Ex_c8659_14515623 0.00209 5B 130.4 wsnp_Ra_c9155_15344108 0.00188 5B 142.5 wsnp_Ra_c20970_30293078 0.00137 5B 155.8 wsnp_Ex_c3422_6283568 0.00132 5D2 34.6 Uncharacterized wsnp_JD_c34368_25929293 0.00136 6A 103.9 uncharacterized wsnp_JD_rep_c48797_33040150 0.00139 6A 103.9 Lr46 Lr17a, Lr17b Uncharacterized Lr16 QTL16,17 Uncharacterized Uncharacterized QTL16,17 Uncharacterized Uncharacterized QTL17 6 wsnp_JD_c3839_4902725 0.00131 6A 111.9 wsnp_BQ167224B_Ta_2_1 0.00148 6B 32.3 Uncharacterized QTL14 wsnp_Ku_c4910_8793327 0.00144 6B 139.8 Uncharacterized QTL17 wsnp_Ex_c5341_9443380 0.00129 7A 53.6 Uncharacterized wsnp_Ex_c5341_9442913 0.00129 7A 53.6 wsnp_Ku_rep_c105954_91953127 0.00141 7A 56.8 wsnp_bq170165A_Ta_1_1 0.00153 7A 97.6 wsnp_Ex_c14248_22204549 0.00131 7B 51.3 wsnp_BE605194B_Ta_2_7 0.00138 7B 105.5 wsnp_BE605194B_Ta_2_1 0.00142 7B 105.7 wsnp_BE498985A_Ta_2_1 0.00156 7B 106.1 wsnp_JD_c13673_13606066 0.00163 7B 132.8 wsnp_Ex_c48087_53105842 0.00077 1A 33.1 Uncharacterized wsnp_Ex_c4092_7395646 0.00094 1B 28.2 Uncharacterized wsnp_Ex_c16581_25100502 0.00096 1B 28.2 wsnp_RFL_Contig1952_1128597 0.00097 1B 28.2 wsnp_Ex_c26860_36084209 0.00153 1B 80.2 wsnp_Ku_c41261_49287074 0.00089 1B 90.7 wsnp_Ex_c2569_4780450 0.00113 1B 91.4 wsnp_Ex_c3147_5817088 0.00105 1B 91.4 wsnp_CAP11_c1043_618449 0.00096 1D 50.8 wsnp_Ku_c53270_57959459 0.00087 1D 67.3 wsnp_CAP11_c1556_864919 0.00098 2A 114.7 wsnp_Ex_c12219_19526749 0.00102 2A 141.0 wsnp_CAP11_c3271_1608092 0.00104 2B 20.5 wsnp_Ku_c2486_4751761 0.00090 2B 23.2 wsnp_Ku_c2486_4751695 0.00093 2B 23.2 wsnp_RFL_Contig3118_3061389 0.00094 2B 23.2 wsnp_BE497494B_Ta_2_1 0.00139 2B 76.0 wsnp_CAP11_c3947_1866837 0.00128 2B 76.0 wsnp_CAP11_c3947_1867089 0.00115 2B 76.0 wsnp_Ex_c62404_62055681 0.00114 2B 76.0 wsnp_Ex_c2430_4546479 0.00115 2B 76.0 wsnp_Ex_c20786_29874875 0.00115 2B 76.4 wsnp_Ex_c20786_29875033 0.00128 2B 76.4 wsnp_Ex_rep_c68704_67559626 0.00129 2B 76.4 wsnp_Ex_c42316_48926687 0.00108 2B 77.5 wsnp_Ex_c12671_20140014 0.00107 2B 77.5 wsnp_Ex_c6537_11338763 0.00095 2B 80.4 wsnp_CAP7_rep_c12606_5316797 0.00094 2B 94.0 wsnp_Ex_c6537_11339130 0.00094 2B 94.0 wsnp_Ex_c41558_48356814 0.00085 2B 158.1 wsnp_Ex_c17700_26446810 0.00089 2B 160.4 wsnp_Ku_c48694_54811376 0.00096 2B 219.9 Sr9b, Sr23 wsnp_Ex_c34303_42642389 0.00101 2B 254.7 Uncharacterized Uncharacterized Lr68 QTL16,18,19 Stem rust Uncharacterized QTL20 Uncharacterized QTL21 Uncharacterized Uncharacterized QTL21,22,23 Uncharacterized Uncharacterized Unpublished QTL22 7 wsnp_Ex_rep_c102478_87635370 0.00106 3A 41.7 wsnp_Ex_rep_c106152_90334299 0.00090 3A 46.2 Uncharacterized wsnp_BF292295A_Ta_2_1 0.00105 3A 46.8 wsnp_Ex_c15036_23203474 0.00107 3A 47.1 wsnp_Ex_c1660_3158776 0.00084 3A 76.0 wsnp_Ra_c7280_12576178 0.00090 3A 81.4 wsnp_Ku_rep_c68861_68030775 0.00086 4A 73.4 Uncharacterized wsnp_BG313770B_Ta_1_1 0.00089 4A 154.3 Uncharacterized wsnp_Ra_c22775_32274079 0.00121 4A 164.3 wsnp_Ex_c49319_53953814 0.00078 4B 61.0 Uncharacterized wsnp_BE497368A_Ta_2_1 0.00109 5A 46.0 Uncharacterized wsnp_BE442892A_Ta_2_2 0.00096 5A 46.0 wsnp_CD453438A_Ta_2_1 0.00090 5A 46.0 wsnp_Ex_c15163_23357477 0.00093 5A 46.0 wsnp_Ku_c39334_47795461 0.00084 6A 10.0 Uncharacterized wsnp_Ex_c14975_23127669 0.00099 6A 78.7 Uncharacterized wsnp_Ex_c99215_85409445 0.00098 6A 78.7 wsnp_CAP11_c3219_1585635 0.00090 6B 0.6 Sr54 wsnp_Ex_c1988_3742022 0.00109 6D 0.7 Uncharacterized wsnp_RFL_Contig2789_2553657 0.00089 7A 41.0 Uncharacterized wsnp_Ra_c31751_40835513 0.00087 7A 74.2 Uncharacterized wsnp_Ku_c4591_8286024 0.00088 7A 98.4 wsnp_CAP11_c827_513472 0.00095 7A 104.9 wsnp_CAP7_c949_486485 0.00089 7A 104.9 wsnp_Ex_c5177_9174930 0.00093 7A 105.5 wsnp_Ex_rep_c68047_66792559 0.00099 7A 118.2 wsnp_CAP8_c334_304253 0.00103 7B 0.0 wsnp_CAP11_c589_398835 0.00100 7B 0.0 wsnp_BM137749D_Ta_2_1 0.00106 7B 133.4 wsnp_Ex_c65899_64135487 0.00091 7D 2.6 Uncharacterized QTL21,24 Uncharacterized Unpublished Uncharacterized The locations of Yr, Lr and Sr genes as per the USDA–ARS Cereal Disease Lab (http://www.ars.usda.gov/Main/docs.htm?docid¼10342) *Unpublished genes were identified through BSA using 9K SNP chip in mapping populations developed from Watkins genotypes and are yet to be named (Bansal and Bariana unpublished results). QTL if reported in the same region are mentioned as well. It may or may not be the same locus. 1 Bariana et al. (2010); 2Lowe et al. (2011); 3Boukaten et al. (2002); 4Carter et al. (2009); 5Mallard et al. (2005); 6Powell et al. (2013); 7Suenaga et al. (2003); 8Singh et al. (2000); 9Prins et al. (2011); 10Lu et al. (2009); 11Yang et al. (2013); 12Hao et al. 2011(2011); 13Lin and Chen (2009); 14Ren et al. 2012(2012); 15 Santra et al. (2008); 16 Messmer et al (2000); 17Chu et al. (2009); 18William et al. (1997); 19Xu et al. (2005); 20Haile et al. (2012); 21Kaur et al. (2009); 22Bhavani et al. (2011); 23Singh et al. (2013), 24Njau et al. (2013) 8 Table S3. P-values and goodness of fit (R2) when regressing genomic prediction accuracy on releationship measures (mean relationship, Gmean; Mean of top X, GtopX). Lr Sr Yr P-value R2 P-value R2 P-value R2 Gtop1 0.000003 0.942709 0.000228 0.833034 0.000025 0.903607 Gtop5 0.000011 0.920800 0.001653 0.729608 0.000672 0.782649 Gtop10 0.000177 0.843265 0.002690 0.695972 0.000158 0.847442 Gtop20 0.000575 0.790723 0.003495 0.676280 0.087855 0.320745 Gtop30 0.013141 0.557183 0.011569 0.570167 0.188512 0.205279 Gtop40 0.085383 0.324880 0.025807 0.482474 0.200709 0.195517 Gtop50 0.037644 0.436212 0.011596 0.569930 0.004206 0.661631 Gmean 0.305028 0.130522 0.127606 0.265318 0.100559 0.300969 Table S4. Accuracy (Pearson correlation) and values for two relationship measures of validation accessions to the reference population: mean genomic relationship (Gmean) and highest genomic relationship (Gtop1). Each accuracy resulted from subsets of low or high relationship in leaf, stem and stripe rust in five repeated random cross-validations each with five folds (30 points per relationship measure). Gmean Trait Cross-Validation Accuracy Gtop1 Accuracy Low High Low High Low High Low High Lr CV1 -0.017 -0.004 0.104 0.362 0.622 1.378 0.027 0.463 Lr CV2 -0.016 -0.003 0.266 0.245 0.596 1.342 0.001 0.492 Lr CV3 -0.015 -0.002 0.302 0.348 0.622 1.367 0.108 0.486 Lr CV4 -0.015 -0.004 0.315 0.191 0.614 1.385 -0.061 0.488 Lr CV5 -0.013 -0.002 0.264 0.299 0.617 1.395 0.116 0.412 Sr CV1 -0.014 -0.003 0.027 0.226 0.612 1.376 -0.038 0.352 Sr CV2 -0.020 -0.001 0.132 0.174 0.629 1.380 0.046 0.253 Sr CV3 -0.013 -0.004 0.219 0.179 0.594 1.349 0.104 0.281 Sr CV4 -0.017 -0.002 0.244 0.261 0.598 1.381 0.147 0.367 Sr CV5 -0.017 -0.001 0.054 0.250 0.594 1.384 0.001 0.288 Yr CV1 -0.022 -0.003 0.468 0.394 0.604 1.351 0.263 0.566 Yr CV2 -0.019 -0.003 0.335 0.520 0.613 1.329 0.368 0.501 Yr CV3 -0.016 -0.002 0.308 0.493 0.608 1.365 0.261 0.507 Yr CV4 -0.015 -0.002 0.242 0.542 0.610 1.368 0.323 0.482 Yr CV5 -0.018 -0.003 0.422 0.444 0.597 1.359 0.295 0.549 Table S5. LogLikelihood and variance components from BLUE models including various random terms, including GxE interactions, where Var(e) is the error variance, Var(ID) is the variance due to accessions, Var(IDxSite) is the variance of accession-by-site interactions, and Var(IDxYear) is the 9 variance of accession-by-year interactions. Models with * are significantly different from the base model that just fitted ID. Random Model Terms LogLikelihood Var(e) Var(ID) Var(IDxSite) Var(IDxYear) ID -1409.5 1.26081 ID IDxSite* -1354.51 1.07312 2.10941 - - 1.53346 0.66856 - ID IDxYear* -1380.14 1.00154 2.0863 - 0.335315 ID IDxSite IDxYear* -1313.34 0.78553 1.45246 0.72856 0.377454 References: Bariana HS, Bansal UK, Schmidt A, Lehmensiek A, Kaur J, Miah H, Howes N, McIntyre CL (2010) Molecular mapping of adult plant stripe rust resistance in wheat and identification of pyramided QTL genotypes. Euphytica 176:251-260 Bhavani S, Singh RP, Argillier O, Huerto-Espino J, Singh S, Njau P (2011) Mapping of durable adult plant stem rust resistance in six CIMMYT wheats to Ug99 group of races. 2011 BGRI technical workshop, St. Paul, Minnesota, USA, pp 43-53 Boukhatem N, Baret PV, Mingeot D, Jacquemin JM (2002) Quantitative trait loci for resistance against Yellow rust in two wheat-derived recombinant inbred line populations. Theoretical and Applied Genetics 104:111-118 Carter A, Chen XM, Garland-Campbell K, Kidwell KK (2009) Identifying QTL for high-temperature adult-plant resistance to stripe rust (Puccinia striiformis f. sp. tritici) in the spring wheat (Triticum aestivum L.) cultivar ‘Louise’. Theoretical and Applied Genetics 119:1119-1128 Chu CG, Friesen TL, Xu SS, Faris JD, Kolmer JA (2009) Identification of novel QTLs for seedling and adult plant leaf rust resistance in a wheat doubled haploid population. Theoretical and Applied Genetics 119:263-269 Haile J, Nachit M, Hammer K, Badebo A, Röder M (2012) QTL mapping of resistance to race Ug99 of Puccinia graminis f. sp. tritici in durum wheat (Triticum durum Desf.). Mol Breeding 30:1479-1493 Hao Y, Chen Z, Wang Y, Bland D, Buck J, Brown-Guedira G, Johnson J (2011) Characterization of a major QTL for adult plant resistance to stripe rust in US soft red winter wheat. Theoretical and Applied Genetics 123:1401-1411 Kaur J, Bansal UK, Khanna R, Saini RG, Bariana HS (2009) Molecular mapping of stem rust resistance in HD2009/WL711 recombinant inbred line population. Int J Plant Breed 3:28-33 Lin F, Chen XM (2009) Quantitative trait loci for non-race-specific, high-temperature adult-plant resistance to stripe rust in wheat cultivar Express. Theoretical and Applied Genetics 118:631-642 Lowe I, Jankuloski L, Chao S, Chen X, See D, Dubcovsky J (2011) Mapping and validation of QTL which confer partial resistance to broadly virulent post-2000 North American races of stripe rust in hexaploid wheat. Theoretical and Applied Genetics 123:143-157 Lu Y, Lan C, Liang S, Zhou X, Liu D, Zhou G, Lu Q, Jing J, Wang M, Xia X, He Z (2009) QTL mapping for adult-plant resistance to stripe rust in Italian common wheat cultivars Libellula and Strampelli. Theoretical and Applied Genetics 119:1349-1359 Mallard S, Gaudet D, Aldeia A, Abelard C, Besnard AL, Sourdille P, Dedryver F (2005) Genetic analysis of durable resistance to yellow rust in bread wheat. Theoretical and Applied Genetics 110:1401-1409 Messmer MM, Seyfarth R, Keller M, Schachermayr G, Winzeler M, Zanetti S, Feuillet C, Keller B (2000) Genetic analysis of durable leaf rust resistance in winter wheat. Theoretical and Applied Genetics 100:419-431 Njau PN, Bhavani S, Huerta-Espino J, Keller B, Singh RP (2013) Identification of QTL associated with durable adult plant resistance to stem rust race Ug99 in wheat cultivar ‘Pavon 76’. Euphytica 190:33-44 Powell NM, Lewis CM, Berry ST, MacCormack R, Boyd LA (2013) Stripe rust resistance genes in the UK winter wheat cultivar Claire. Theoretical and Applied Genetics 126:1599-1612 Prins R, Pretorius ZA, Bender CM, Lehmensiek A (2011) QTL mapping of stripe, leaf and stem rust resistance genes in a Kariega × Avocet S doubled haploid wheat population. Mol Breeding 27:259-270 10 Ren Y, Li Z, He Z, Wu L, Bai B, Lan C, Wang C, Zhou G, Zhu H, Xia X (2012) QTL mapping of adult-plant resistances to stripe rust and leaf rust in Chinese wheat cultivar Bainong 64. Theoretical and Applied Genetics 125:1253-1262 Santra DK, Chen XM, Santra M, Campbell KG, Kidwell KK (2008) Identification and mapping QTL for high-temperature adult-plant resistance to stripe rust in winter wheat (Triticum aestivum L.) cultivar ‘Stephens’. Theoretical and Applied Genetics 117:793-802 Singh RP, Nelson JC, Sorrells ME (2000) Mapping Yr28 and Other Genes for Resistance to Stripe Rust in Wheat. Crop Sci 40:1148-1155 Singh S, Singh R, Bhavani S, Huerta-Espino J, Eugenio L-V (2013) QTL mapping of slow-rusting, adult plant resistance to race Ug99 of stem rust fungus in PBW343/Muu RIL population. Theoretical and Applied Genetics 126:1367-1375 Suenaga K, Singh RP, Huerta-Espino J, William HM (2003) Microsatellite Markers for Genes Lr34/Yr18 and Other Quantitative Trait Loci for Leaf Rust and Stripe Rust Resistance in Bread Wheat. Phytopathology 93:881-890 William HM, Hoisington D, Singh RP, Gonzalez-de-Leon D (1997) Detection of quantitative trait loci associated with leaf rust resistance in bread wheat. Genome 40:253-260 Xu X, Bai G, Carver BF, Shaner GE, Hunger RM (2005) Molecular Characterization of Slow LeafRusting Resistance in Wheat A portion of this research was funded by the Oklahoma Wheat Research Foundation and the Oklahoma Agric. Exp. Stn. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. Crop Sci 45:758-765 Yang EN, Rosewarne GM, Herrera-Foessel SA, Huerta-Espino J, Tang ZX, Sun CF, Ren ZL, Singh RP (2013) QTL analysis of the spring wheat “Chapio” identifies stable stripe rust resistance despite intercontinental genotype × environment interactions. Theoretical and Applied Genetics 126:1721-1732 11