Electronic Supplemental Material
advertisement

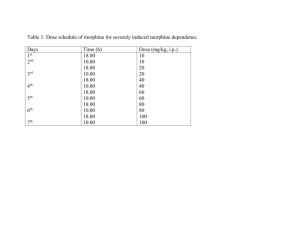
Electronic Supplemental Material Morphine and Metabolites in Young Children: a Population Pharmacokinetic Model Authors: Nieves Velez de Mendizabal*1,2, Ricardo Jimenez-Mendez*3, 4, 5, Erin Cooke6, Carolyne J. Montgomery6, Joy Dawes6, Michael J. Rieder7, Katarina Aleksa8, Gideon Koren8, Carlos O. JacoboCabral9, Rodrigo Gonzalez-Ramirez9, Gilberto Castañeda-Hernandez9 and Bruce C. Carleton3,4,5 Affiliations: 1 Division of Clinical Pharmacology, Department of Medicine, Indiana University School of Medicine, Indianapolis 2 Indiana Clinical and Translational Sciences Institute (CTSI), Indianapolis, IN, USA 3 Division of Translational Therapeutics, Department of Paediatrics, Faculty of Medicine, University of British Columbia, Vancouver, Canada 4 Pharmaceutical Outcomes Programme, BC Children’s Hospital, Vancouver, Canada 5 Child & Family Research Institute, Vancouver, Canada 6 Pediatric Anesthesia Research Team, University of British Columbia and Department of Pediatric Anesthesia, BC Children's Hospital, Vancouver, Canada 7 Department of Physiology and Pharmacology, Western University, London, Canada 8 The Motherisk Program, Division of Clinical Pharmacology/Toxicology, The Hospital for Sick Children Toronto, Canada 9 Departamento de Farmacología, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, México, DF. Corresponding author: Dr. Bruce C. Carleton Child and Family Research Institute 950 West 28th Avenue, Rm. A3-207 Vancouver, BC. V5Z 4H4, Canada e-mail: bcarleton@popi.ubc.ca * Nieves Velez de Mendizabal and Ricardo Jimenez-Mendez contributed equally to this work. Running Title: PK model for morphine and its metabolites Keywords: Morphine, M3G, M6G, NONMEM, children Abbreviations: M3G, morphine-3-glucoronide; M6G, morphine-6-glucoronide; IV, intravenous; ISV, inter-subject variability; CWRES, conditional weighted residuals; NPDE, normalized prediction distribution errors; RSE, relative standard error; CV, coefficient of variation; PK, pharmacokinetics 1 Online Resource 1: Bio-analytical assay After induction of anaesthesia, a second IV cannula was inserted. Blood samples were drawn at 30, 60, 90, 120, 180 and 240 minutes. Blood samples were collected in heparinized tube and refrigerated. Samples were centrifuged at 3000 bpm and stored at -80 °C until plasma concentration of morphine, M6G and M3G were measured using a validated high performance liquid chromatography essay. Sample preparation summary: Morphine and morphine glucuronides (morphine-6-glucuronide (M-6-G) and morphine-3-glucuronide (M-3-G)) were extracted from plasma by solid-phase extraction on C18 cartridges at pH 9.3. Method from Leis HJ et al., (2002) LC-MS/MS summary Analysis was performed on an ABSciex QTRAP5500 (ABSciex: Framingham, Massachusetts, USA) and Agilent 1290 LC system (Agilent Technologies: Santa Clara, California, USA). Chromatography ran at a flow rate of 800 µL/min on a Kinetex HILIC column 4.6x50mm, 2.6 µm (Agilent) with a gradient starting at 5% A (90/10 Water/Acetonitrile 5 mM ammonium Formate pH 3.2) and 95% B (10/90 Water/Acetonitrile 5 mM ammonium Formate pH 3.2) ramping to 100% A at 5 minutes. This method allowed for baseline chromatographic resolution of the 2 glucuronide isomers. The mass spectrometer was operated in positive ESI mode with a source temperature of 650°C and an IS Voltage setting of 5000. Precursor to product ion mass transitions were established by standard infusions. Data was acquired by MRM mode (Multiple Reaction Monitoring) with mass transitions as follows: 286→ 165 m/z for Morphine, 289 → 165 m/z for Morphine-d3 (RT 3.0 min) 462 → 286 m/z for Morph-6-Gluc, 465 → 289 for Morph-6-Gluc-d3 (RT 4.85 min) 462 → 286 m/z for Morph-3-Gluc, 465 → 289 for Morph-3-Gluc-d3 (RT. 5.05 min) Data analysis and peak integration was performed using Analyst 1.5.2 software from ABSciex. Sample concentrations were calculated by plotting peak area ratios (Analyte/Internal Standard) against calibration curves of extracted matrix spiked standards. Quality control follow-up Sample concentrations were calculated by plotting peak area ratios (Analyte/Internal Standard) against calibration curves of extracted matrix spiked standards Linearity was verified by presenting regression line coefficients (r2). The Acceptance Criteria of the r2 of the regression line was > 0.99 Quality controls of extracted matrix spiked standards were run along with the unknown samples, and their concentrations were calculated by plotting peak area ratios (Analyte/Internal Standard) against calibration curves of extracted matrix spiked standards. Percent recovery was >80% and <120%. 2 Online Resource 2: Ordinary Differential Equations ODEs of the selected PK model for morphine and its metabolites M3G and M6G dA1 A1 (t) k a dt dA 2 CL EX k A1 (t) k a A 2 (t) A 2 (t) m dt VC VC dA 3 CL 2M3G CL M3G A 5 (t) A 3 (t) dt VC VM3 CL M6G dA 4 CL 2M6G A 5 (t) A 4 (t) dt VC VM6 dA 5 CL 2M3G CL 2M6G k A 2 (t) m A 5 (t) A 5 (t) dt VC VC VC where A1(t), A2(t), A3(t), A4(t) and A5(t) represent depot, morphine central, metabolite M3G central, metabolite M6G central and delay transit compartment respectively; ka first order rate constant of absorption; VC/F is apparent volume of distribution of the central compartment; kmf is the first order rate constant for the metabolism transit compartment, CL2M3G and CL2M6G are the formation rates for M3G and M6G respectively and CLM3G and CLM6G are the elimination rates. The metabolite volumes of distribution (V3M, V6M) were parameterized as VC/F. 3 Online Resource 3: NONMEM model $SUBROUTINES ADVAN13 TOL=9 ;TIME (MINUTES) $MODEL COMP(DEPOT) COMP(PARENT) ; 2 CENTRAL MORFINA COMP(M3G) ; 3 METABOLITO M-3-G COMP(M6G) ; 4 METABOLITO M-6-G COMP(METAB) ; 5 LIVER METABOLISM $PK KA = THETA(1)*EXP(ETA(1)) ; KA (min-1) VC = THETA(2)*EXP(ETA(2)) ; V CENTRAL MORPHINE (L) CLEX = THETA(3)*EXP(ETA(3)) ; CLEX MORPHINE-ELIMINATION (L/min) F1 = THETA(4)*EXP(ETA(4)) ; Bioavailability CL2M3G = THETA(5)*EXP(ETA(5)) ; FORMATION DE M-3-G CLM3G = THETA(6)*EXP(ETA(6)) ; ELIMINATION DE M-3-G CL2M6G=THETA(7)*EXP(ETA(7)) ; FORMATION DE M-6-G CLM6G=THETA(8)*EXP(ETA(8)) ; ELIMINATION DE M-6-G KMF=THETA(9)*EXP(ETA(9)) ; METABOLISM (DELAY) VM3=VC ; V MORPHINE-3-GLUCURONIDO VM6=VC ; V MORPHINE-6-GLUCURONIDO ;-------- scales -------S2=VC/1000 ; AMT= mg, DV= ng/ml S3=VM3G/1000 ; AMT= mg, DV= ng/ml S4=VM6G/1000 ; AMT= mg, DV= ng/ml $DES DADT(1)=-KA*A(1) DADT(2)=KA*A(1)-(CLEX/VC)*A(2)-(KMF/VC)*A(2) ;MORPHINE DADT(3)=(CL2M3G/VC)*A(5)-(CLM3G/VM3)*A(3) ;M-3-G DADT(4)=(CL2M6G/VC)*A(5)-(CLM6G/VM6)*A(4) ;M-6-G DADT(5)=(KMF/VC)*A(2) -(CL2M3G/VC)*A(5) - (CL2M6G/VC)*A(5) … 4 Online Resource 4: Individual and population predictions versus observations Fig. S1 Individual and population predictions versus observations. Dots stand for observations, solid lines for the model individual predictions and dashed red lines for population predictions. Grey color represents morphine, blue M3G and green M6G concentrations. Results are stratified by dose: (A) 0.1 mg/kg; (B) 0.2 mg/kg and (C) 0.3 mg/kg. 5