Supporting Information
advertisement
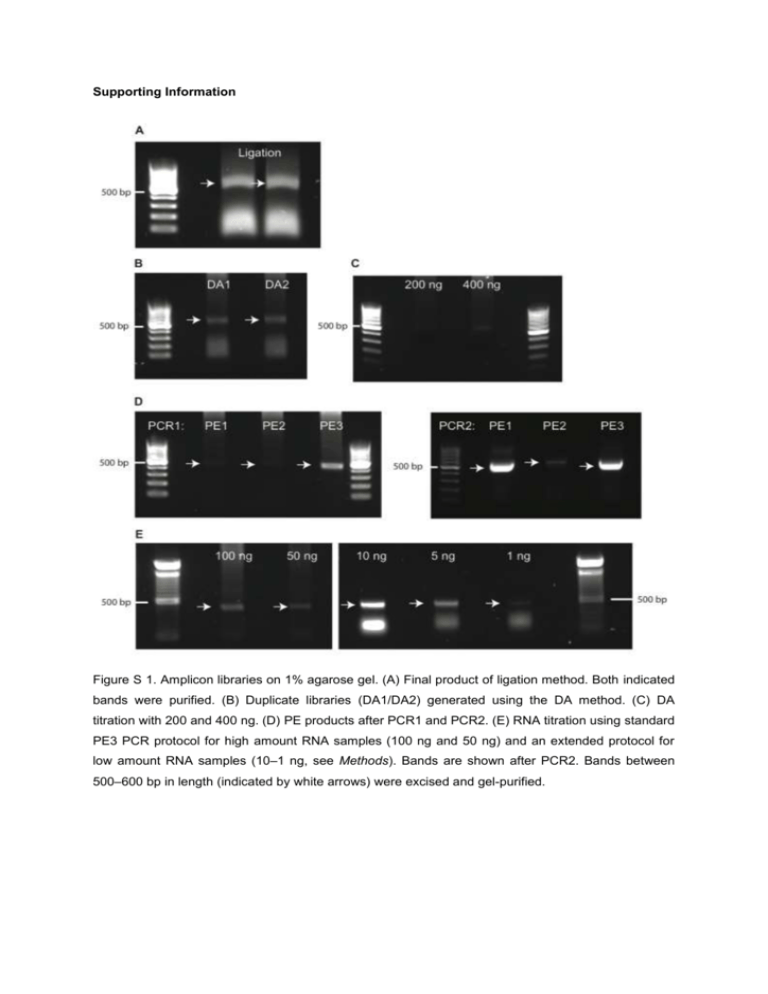
Supporting Information Figure S 1. Amplicon libraries on 1% agarose gel. (A) Final product of ligation method. Both indicated bands were purified. (B) Duplicate libraries (DA1/DA2) generated using the DA method. (C) DA titration with 200 and 400 ng. (D) PE products after PCR1 and PCR2. (E) RNA titration using standard PE3 PCR protocol for high amount RNA samples (100 ng and 50 ng) and an extended protocol for low amount RNA samples (10–1 ng, see Methods). Bands are shown after PCR2. Bands between 500–600 bp in length (indicated by white arrows) were excised and gel-purified. B 95 10 10-2 CDR3 frequency [%] 25 Reliably detected CDR3s: 12,069 100 95 75 25 10-3 Frequency range : 1.7 - 3.3 ´ 10-4 Abundance range: 25,536–5 Replicate DA1 Replicate DA2 97% 97.1% 100 75 50 25 0 D 14000 10 8000 10000 12000 14000 12000 10000 8000 10-1 10-2 10 -3 Frequency range : 1.7 - 2.9 ´ 10 DA2 8000 6000 4000 2000 2000 4000 6000 8000 14000 8000 10000 12000 14000 12000 -4 10000 Abundance range: 23,135–4 0 C 0 Replicate DA2 Replicate DA2 50 Replicate DA1 50 0 10-1 Replicate DA1 Unique CDR3s present in all compared datasets [%] 75 0 Reads mapping to reliably detected CDR3s [%] A 100 8000 DA1 r=0.93 8000 p<0.01 6000 Reliably detected CDR3s: 12,318 4000 100 101 102 103 Number of unique CDR3s 104 100 101 102 103 Number of unique CDR3s 104 2000 2000 4000 6000 8000 0 5000 Figure S 2. DA duplicates (DA1/DA2, technical replicates) yield highly comparable HTS datasets. (A) Reliably detected CDR3s were determined as described in Fig. 2 using a 95% reliable detection cutoff. On average, ≈12,200 CDR3s were reliably detected. (B) Their corresponding average frequency and abundance ranges are shown (≈1.7–3.3x10-4% and ≈24,000–5 reads per unique CDR3, respectively). (C) Reliably detected CDR3s corresponded to ≈97% of total sequencing reads. (D) The Spearman rank correlation coefficient was high (r=0.93). B 95 10 10-1 50 0 10-2 CDR3 frequency [%] 25 Reliably detected CDR3s: 11,661 100 95 75 DA DA 10-3 Frequency range : 1.7 - 3.3 ´ 10-4 Abundance range: 25,536–5 C DA PE 96.9% 95.7% 100 75 50 25 0 D 8000 10000 12000 10 0 12000 10000 8000 PE 10-1 6000 4000 50 PE PE Unique CDR3s present in all compared datasets [%] 75 0 Reads mapping to reliably detected CDR3s [%] A 100 10-2 2000 2000 4000 6000 8000 10000 12000 25 10 -3 12000 Frequency range : 1.2 - 4.5 ´ 10-4 10000 Abundance range: 15,380–6 DA 0 Reliably detected CDR3s: 11,297 100 101 102 103 Number of unique CDR3s r=0.9 8000 p<0.01 6000 4000 104 100 101 102 103 Number of unique CDR3s 104 2000 2000 4000 6000 2000 4000 6000 8000 Figure S 3. Pairwise comparison of HTS datasets from antibody repertoire libraries prepared using the DA and PE method. (A) Reliably detected CDR3s were determined as described in Fig. 2 using a 95% reliable detection cut-off. On average, ≈11,500 CDR3s were reliably detected. (B) Their corresponding average frequency and abundance ranges are shown (≈1.5–3.9x10-4 % and ≈20,000–5 reads per unique CDR3, respectively). (C) Reliably detected CDR3s corresponded to ≈96% of total sequencing reads. (D) The Spearman rank correlation coefficient was high (r=0.90). B 95 10 10-1 50 10-2 Frequency range : 1.9 - 8.2 ´ 10 0 CDR3 frequency [%] 25 Reliably detected CDR3s: 8,831 100 95 75 Ligation Ligation 10-3 -4 Abundance range: 9,254–4 C Ligation DA 96.5% 95.4% 100 75 50 25 0 D 10000 10 6000 6000 4000 10-2 10 -3 2000 2000 10000 6000 8000 r=0.86 6000 102 103 p<0.01 4000 Reliably detected CDR3s: 8,570 101 6000 10000 Ligation Number of unique CDR3s 4000 8000 Abundance range: 25,536–12 6000 100 10000 DA Frequency range : 1.7 - 7.9 ´ 10-4 25 8000 8000 DA 50 0 6000 0 10-1 DA Unique CDR3s present in all compared datasets [%] 75 0 Reads mapping to reliably detected CDR3s [%] A 100 104 100 101 102 103 Number of unique CDR3s 104 2000 2000 4000 6000 2000 4000 6000 Figure S 4. Pairwise comparison of HTS datasets from antibody repertoire libraries prepared using the ligation and DA method. (A) Reliably detected CDR3s were determined as described in Fig. 2 using a 95% reliable detection cut-off. On average, ≈8,700 CDR3s were reliably detected. (B) Their corresponding average frequency and abundance ranges are shown (≈1.8–8.1x10-4% and ≈17,000–8 reads per unique CDR3, respectively). (C) Reliably detected CDR3s corresponded to ≈96% of total sequencing reads. (D) The Spearman rank correlation coefficient was high (r=0.86). B 95 10 10-1 50 10-2 Frequency range : 1.9 - 6.1 ´ 10-4 CDR3 frequency [%] 25 0 Ligation Ligation Reliably detected CDR3s: 9,111 100 95 75 10 -3 Abundance range: 9,254–3 C Ligation PE 96.7% 93.3% 100 75 50 25 0 D 10000 10 6000 10-2 PE Frequency range : 1.2 - 1.3 ´ 10 10000 6000 8000 102 103 6000 10000 r=0.79 6000 p<0.01 4000 Reliably detected CDR3s: 7,925 101 4000 8000 Ligation Number of unique CDR3s 6000 2000 2000 -3 10-3 100 10000 4000 Abundance range: 15,380–17 25 8000 8000 PE 50 0 6000 0 10-1 PE Unique CDR3s present in all compared datasets [%] 75 0 Reads mapping to reliably detected CDR3s [%] A 100 104 100 101 102 103 Number of unique CDR3s 104 2000 0 2000 4000 2000 4000 6000 0 Figure S 5. Pairwise comparison of HTS datasets from antibody repertoire libraries using the ligation and PE method. A) Reliably detected CDR3s were determined as described in Fig. 2 using a 95% CDR3 reliable detection cut-off. On average, ≈8,700 CDR3s were reliably detected. (B) Their corresponding average frequency and abundance ranges are shown (≈1.6–3.7x10-4% and ≈12,500– 10 per unique CDR3, respectively). (C) Reliably detected CDR3s corresponded to ≈95% of total sequencing reads. (D) The Spearman rank correlation coefficient was high (r=0.79). B 100 10-1 75 10-3 100 95 25 0 10-1 100 75 10-1 Frequency range : 1.1 - 2.6 ´ 10 Abundance range: 24,670–6 50 10 10 10 10 Number of unique CDR3s 96.3% 0 10 4 10 10000 15000 10000 10000 5000 5000 10000 15000 10000 Frequency range : 1.2 - 3 ´ 10 Abundance range: 15,380–4 10 0 10 1 10 2 -4 10 3 15000 PE3 5000 15000 10 Number of unique CDR3s PE2 10000 15000 r=0.93 10000 p<0.01 5000 10000 3 95.9% 25 10-4 Reliably detected CDR3s: 13,167 2 96.2% 75 -4 -2 10-3 25 1 PE3 PE3 PE3 50 0 PE2 10-4 Reliably detected CDR3s: 13,013 100 95 0 PE1 100 D 10-2 10-3 C PE2 50 CDR3 frequency [%] 10 75 0 Frequency range : 1.1 - 2.6 ´ 10-4 Abundance range: 20,939–5 10-4 Reliably detected CDR3s: 12,948 PE2 Unique CDR3s present in all compared datasets [%] 10-2 25 0 PE1 PE1 50 Reads mapping to reliably detected CDR3s [%] A 100 95 10000 PE1 10000 5000 10000 15000 10000 r=0.94 r=0.93 p<0.01 p<0.01 5000 4 5000 10000 5000 10000 5000 10000 Figure S 6. Variations in the PE protocol have minimal effects on HTS datasets. PE1 and PE2 were prepared with 8 cycles in the last annealing temperature step of PCR1 (16 total cycles) and either amplified with 12 (PE1) or 8 (PE2) total cycles in PCR2, resulting in an overall number of 28 and 24 cycles, respectively. For PE3, 4 cycles were switched from PCR2 to PCR1, having 12 cycles in the last annealing temperature step of PCR1 (20 total cycles) and a total of 8 cycles in PCR2, resulting in an overall cycle number of 28 (equal cycle number to PE1 and standard conditions used for ligation and DA). (A) Reliably detected CDR3s were determined as described in Fig. 2 using a 95% CDR3 reliable detection cut-off. On average, ≈13,000 CDR3s were reliably detected. (B) Their corresponding average frequency and abundance ranges are shown (≈1.1–2.7x10-4% and ≈20,000–5 per unique CDR3, respectively). (C) Reliably detected CDR3s corresponded to ≈96% of sequencing reads. (D) The Spearman rank correlation coefficients were high (r≈0.93). A B 100 95 10 10 10 -2 0 -1 100 ng 100 ng 50 10-2 10-3 25 Frequency range : 1.1 - 2.3 ´ 10-2 Abundance range: 13,000–281 Reliably detected CDR3s: 837 100 ng 50 ng 10 ng 82.5% 50 25 46.3% 48.8% 49.1% 44.9% 44.2% Reliably detected CDR3s: 835 75 10-3 Abundance range: 10,771–213 D 2500 100 1500 2000 2500 2000 1500 10 -1 10-2 10-3 25 Frequency range : 1.2 - 2.3 ´ 10-2 1500 Frequency range : 1.1 - 2.7 ´ 10-2 1500 1000 1500 100 10 ng1500 10 -2 10-3 25 5 ng 5 ng 50 1500 50 ng1500 1000 1500 100 100 ng1500 10-2 10-3 25 1 ng 1 ng 50 Frequency range : 1.2 - 1.2 ´ 10-2 1500 1500 1500 1000 1000 1000 p<0.01 1000 1500 p<0.01 0 500 r=0.77 p<0.01 1000 1500 1000 r=0.41 p<0.01 1000 r=0.84 p<0.01 1000 r=0.92 500 ng1500 1000 1000 r=0.39 1500 p<0.01 0 500 1500 r=0.78 1000 p<0.01 1000 1500 r=0.39 p<0.01 1000 1500 p<0.01 0 500 0 500 1000 2000 2000 1000 r=0.91 p<0.01 1500 Abundance range: 8,197–85 1500 r=0.92 1500 1000 500 2000 2000 10-1 75 1500 r=0.84 1500 Frequency range : 1 - 2.9 ´ 10-2 p<0.01 0 2000 2000 Abundance range: 18,636–535 Reliably detected CDR3s: 629 1000 r=0.76 1500 1000 r=0.38 5 ng 1500 1500 2000 2000 10-1 75 500 2000 2000 Abundance range: 17,175–445 Reliably detected CDR3s: 676 1500 1 ng 1000 500 1000 1500 10 ng 10 ng 50 10-2 CDR3 frequency [%] 25 0 50 ng 50 0 1 ng 75 10-1 75 0 100 95 5 ng 100 50 ng Unique CDR3s present in all compared datasets [%] Frequency range : 1.2 - 2.4 ´ 10 Abundance range: 15,380–320 75 0 100 95 -3 Reliably detected CDR3s: 767 Reads mapping to reliably detected CDR3s [%] 10-2 25 0 100 95 500 ng 500 ng 50 0 100 95 500 ng 100 10-1 75 0 100 95 C 100 1000 1500 r=0.85 p<0.01 1000 1500 r=0.78 p<0.01 1000 1500 r=0.41 p<0.01 1000 1500 p<0.01 1000 Reliably detected CDR3s: 2,189 0 10 1 10 2 10 10 3 Number of unique CDR3s 10 4 10 0 10 1 10 2 10 3 10 4 Number of unique CDR3s Figure S 7. Comparison of HTS datasets from antibody repertoire libraries prepared using the PE method and lowering the amounts of total RNA input (500–1 ng). Including the 1 ng dataset into the comparison of 500–5 ng datasets reduced the number of reliably detected CDR3s and CDR3 rank correlation (Fig. 4, Table S1). (A) Reliably detected CDR3s were determined as described in Fig. 2 using a 95% reliable detection cut-off. On average, ≈1,000 CDR3s were reliably detected. (B) Their corresponding average frequency and abundance ranges are shown (≈1.1–2.3x10-2% and ≈14,000– 300 per unique CDR3, respectively). (C) Reliably detected CDR3s corresponded to ≈52% of total sequencing reads. (D) The Spearman rank correlation coefficients ranged between r=0.92 and r=0.38. B 95 10-1 10-2 Frequency range : 5.5 ´ 10-1 - 5.7 ´ 10-4 VDJ frequency [%] 25 Reliably detected VDJs: 31,343 0 100 95 50 10 -3 Abundance range: 4,772–5 82.5% 50 25 0 D 30000 10-1 20000 Frequency range : 5.7 ´ 10-1 - 5.3 ´ 10-4 10000 10000 30000 101 102 40000 30000 Abundance range: 4,304–4 3 10 10 4 100 105 101 102 103 104 10 5 100 100 95 75 DA1 trimmed 10-1 DA1 trimmed 50 10-2 Frequency range : 1.5 - 4.7 ´ 10-4 VDJ frequency [%] 25 Reliably detected VDJs: 13,267 100 95 10-3 25 0 10000 20000 DA1 trimmed DA2 trimmed 87.3% 87.9% 100 75 50 25 0 H 10000 15000 15000 10000 10000 DA2 trimmed 10 -1 DA2 trimmed DA2 trimmed 50 Abundance range: 15,952–5 20000 G 100 75 10-2 Frequency range : 1.5 - 5.4 ´ 10-4 10-3 Abundance range: 14,380–5 5000 5000 10000 15000 10000 10000 15000 r=0.9 10000 DA1 trimmed Reliably detected VDJs: 13,726 102 p<0.01 10000 10000 Number of unique VDJs F 101 r=0.68 DA1 untrimmed 20000 E 100 20000 40000 Reads mapping to reliably detected VDJs [%] 0 40000 DA2 untrimmed Number of unique VDJs Unique VDJs present in all compared datasets [%] 76.9% 75 30000 10-3 10 0 DA2 untrimmed Reliably detected VDJs: 39,416 0 0 DA1 untrimmed 40000 10-2 25 C 100 DA2 untrimmed 75 DA2 untrimmed Unique VDJs present in all compared datasets [%] 50 DA1 untrimmed DA1 untrimmed 75 Reads mapping to reliably detected VDJs [%] A 100 p<0.01 5000 103 Number of unique VDJs 104 105 100 101 102 103 104 10 5 Number of unique VDJs 5000 10000 5000 10000 Figure S 8. VDJ primer trimming reduces VDJ diversity. The effect of primer trimming on VDJ diversity was assessed using the DA method duplicate (DA1/DA2). Primer trimming of VDJ regions was performed as detailed in Methods. (A/E) Reliable detected VDJs were determined as described in Fig. 2 using a 95% VDJ reliable detection cut-off. On average, ≈35,400 and ≈13,500 VDJs were reliably detected for non-trimmed and trimmed HTS datasets, respectively. (B/F) Their corresponding average frequency and abundance ranges are shown. (C/G) Reliably detected VDJs corresponded to ≈80/≈88% of total sequencing reads. (D/H) The Spearman’s rank correlation coefficients were r=0.68 and r=0.90 for non-trimmed and trimmed HTS datasets, respectively. Table S 1. RNA titration (500–5 ng) using the PE method. On average, ≈6,200 CDR3s were reliably detected, which extended over average frequency and abundance ranges of ≈1.1–2.5x10-3% and ≈15,000–34 reads per unique CDR3, respectively. Reliably detected CDR3s corresponded to ≈92% of total sequencing reads. The corresponding Spearman rank correlation coefficients are shown in Fig. 4. HTS dataset (PE) 500 ng 100 ng 50 ng 10 ng 5 ng Average Number of reliably detected CDR3s Frequency range [%] Abundance range 6,363 6,327 6,333 5,858 6,155 6,207 1.2–2.4x10-3 1.1–2.5x10-3 1.2–2.5x10-3 1.1–2.7x10-3 1.0–2.2x10-3 15,380–32 13,000–30 10,771–23 17,175–44 18,636–40 14,992–34 1.1–2.5x10-3 Reads mapping to reliably detected CDR3s [%] 93.4 91.0 92.4 92.1 90.5 91.9 Table S 2. The 30 highest ranked CDR3 amino acid sequences of all RNA titration datasets are shown in a color-coded manner with respective frequencies. Color code based on CDR3s from the 500 ng sample: Top 1–5: bright fill, white font; Top 6–10: bright fill, black font; Top 11–15: Dark fill, white font; Top 16– 20: grey fill, white font; Top 21–25: pastel fill, black font; Top 26–30: yellow fill, black font. Clones present in 5 out of 6 datasets are marked in red font. Clones, present in less than five out of the six datasets are not color-indicated. The mean Spearman rank correlation coefficient of datasets 500–1ng is r=0.81. Freq.: frequency of clones in percent. 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 500 ng Sequence ARWEYYAMDY ARLNYYGNLFV ARHAYYDQTEVSFVY ARLDLFAY ARHHRYAYYFDY ASYRYGWYFDV ARDVGYGNYFVY ARSGKVRNAMDY ARYRLGTYFDY ASYVYGYYFDY TREGDSPFAY ATYDGYYFDY ARLIYGSSYIVDF ARGGISVDY TTGFAY AAYYYGSSYGFAY ARLGYYGWFAY ARSGSSSGYFDY AREARTTARFAY ARLGYYGYFAY ARLGYYGGFAY ARDKGRVFFDY ATYDGYCFDY ARWELGRAWFAY ARDYGSSHFDH ARSGGYGNYLAWFAY ARFPMDY ARGGHDFSLDY SRHYYSTPYYAMDY ARDSFLITRALDY Freq. 1.156 0.802 0.528 0.449 0.433 0.420 0.388 0.342 0.296 0.288 0.261 0.254 0.251 0.250 0.226 0.225 0.222 0.220 0.215 0.215 0.207 0.203 0.200 0.193 0.186 0.183 0.182 0.182 0.179 0.171 100 ng Sequence ARWEYYAMDY ARLNYYGNLFV ARHAYYDQTEVSFVY ARHHRYAYYFDY ARDVGYGNYFVY ASYRYGWYFDV ARLDLFAY ARSGKVRNAMDY ATYDGYYFDY ARYRLGTYFDY ASYVYGYYFDY ARLIYGSSYIVDF ARLGYYGWFAY ARGGISVDY ARSGSSSGYFDY ARLGYYGYFAY AREARTTARFAY ARLGYYGGFAY TREGDSPFAY ARDKGRVFFDY ATYDGYCFDY TTGFAY AAYYYGSSYGFAY TRGGNWEDFDY ARFPMDY SRHYYSTPYYAMDY ARDSFLITRALDY ARGGHDFSLDY ARWELGRAWFAY ARSGGYGNYLAWFAY Freq. 1.070 0.890 0.630 0.474 0.443 0.410 0.403 0.341 0.288 0.286 0.268 0.253 0.252 0.249 0.249 0.249 0.242 0.238 0.231 0.226 0.224 0.217 0.215 0.212 0.194 0.192 0.190 0.189 0.187 0.183 50 ng Sequence ARWEYYAMDY ARLNYYGNLFV ARHAYYDQTEVSFVY ARDVGYGNYFVY ARLDLFAY ARHHRYAYYFDY ASYRYGWYFDV ARSGKVRNAMDY ATYDGYYFDY ARYRLGTYFDY ASYVYGYYFDY ARGGISVDY ARLGYYGWFAY ARLGYYGYFAY ARSGSSSGYFDY TTGFAY ARLGYYGGFAY ARLIYGSSYIVDF ATYDGYCFDY AREARTTARFAY TREGDSPFAY ARDKGRVFFDY ARLDYYGYLNY AAYYYGSSYGFAY ARFPMDY TRGGNWEDFDY ARWELGRAWFAY ARLSYYGRFAY AKNYYYGYVAY ARQGYYGKGDY Freq. 1.153 0.927 0.594 0.467 0.463 0.457 0.374 0.348 0.332 0.311 0.289 0.273 0.267 0.262 0.258 0.250 0.246 0.241 0.229 0.228 0.228 0.218 0.211 0.209 0.207 0.198 0.192 0.191 0.185 0.185 10 ng Sequence ARWEYYAMDY ARLNYYGNLFV ARHAYYDQTEVSFVY ARHHRYAYYFDY ARLDLFAY ARDVGYGNYFVY ASYRYGWYFDV ARSGKVRNAMDY ARLIYGSSYIVDF ATYDGYYFDY ASYVYGYYFDY ARGGISVDY ARLGYYGWFAY ARYRLGTYFDY TTGFAY AREARTTARFAY ATYDGYCFDY ARLGYYGYFAY ARSGSSSGYFDY TRGGNWEDFDY AAYYYGSSYGFAY ARLGYYGGFAY ARWELGRAWFAY TREGDSPFAY ARDSFLITRALDY ARDKGRVFFDY ARLDYYGYLNY ARFPMDY AKNYYYGYVAY SRHYYSTPYYAMDY Freq. 1.055 0.811 0.576 0.518 0.449 0.426 0.354 0.314 0.276 0.276 0.263 0.263 0.262 0.262 0.253 0.252 0.252 0.244 0.242 0.231 0.213 0.209 0.202 0.202 0.200 0.191 0.191 0.188 0.183 0.182 5 ng Sequence ARWEYYAMDY ARLNYYGNLFV ARHAYYDQTEVSFVY ARHHRYAYYFDY ARDVGYGNYFVY ARLDLFAY ASYRYGWYFDV ARSGKVRNAMDY ARWELGRAWFAY ARSGSSSGYFDY ATYDGYYFDY ARDINYDIYWYFDV ARGGISVDY ASYVYGYYFDY ARYRLGTYFDY ARLGYYGGFAY ARLGYYGYFAY TRGGNWEDFDY ARLGYYGWFAY TREGDSPFAY ATYDGYCFDY ARDYGSSHFDH TTGFAY ASYRYAWYFDV ARLIYGSSYIVDF ARFPMDY AREARTTARFAY ARSGGYGNYLAWFAY ARDKGRVFFDY ARLDYYGYLNY Freq. 1.021 0.916 0.635 0.497 0.415 0.414 0.391 0.319 0.277 0.262 0.254 0.250 0.248 0.239 0.235 0.235 0.232 0.222 0.219 0.217 0.215 0.209 0.205 0.204 0.204 0.202 0.201 0.201 0.198 0.195 1ng Sequence ARWEYYAMDY ARLNYYGNLFV ARHAYYDQTEVSFVY ARHHRYAYYFDY ASYRYGWYFDV ARDVGYGNYFVY ATYDGYYFDY AAYYYGSSYGFAY ATYDGYCFDY ASYVYGYYFDY ARGGISVDY ARYRLGTYFDY ARDINYDIYWYFDV ANYYYGSSLFAY ARLDLFAY ARLIYGSSYIVDF ARLGYYGYFAY ARLGYYGGFAY ARDKGRVFFDY TTGFAY ARLGYYGWFAY ARSGKVRNAMDY ARGGANVDY AREARTTARFAY ARKGAMITHYYAMDY TIGGFDY TREGTYDGCSGHFDY ARGGHDFSLDY ARWELGRAWFAY ARHGITRLLDY Freq. 1.187 0.776 0.739 0.565 0.532 0.440 0.392 0.338 0.316 0.311 0.302 0.287 0.279 0.278 0.273 0.268 0.253 0.250 0.239 0.236 0.228 0.224 0.222 0.219 0.217 0.215 0.214 0.214 0.197 0.196 Table S 3. List of all primers used for ligation, DA, and PE. fw: forward, rv: reverse, UAd: universal adapter, Idx: index, RC: reverse complement. Ligation 1 DA IgH Universal Forward TruSeq Universal Adapter + Diversity region + VH 5' specific region IgH-UAd-fw1 IgH-UAd-fw2 IgH-UAd-fw3 IgH-UAd-fw4 IgH-UAd-fw5 IgH-UAd-fw6 IgH-UAd-fw7 IgH-UAd-fw8 IgH-UAd-fw9 IgH-UAd-fw10 IgH-UAd-fw11 IgH-UAd-fw12 IgH-UAd-fw13 IgH-UAd-fw14 IgH-UAd-fw15 IgH-UAd-fw16 IgH-UAd-fw17 IgH-UAd-fw18 IgH-UAd-fw19 Universal Adapter - NNNN GAKGTRMAGCTTCAGGAGTC Universal Adapter - NNNN GAGGTBCAGCTBCAGCAGTC Universal Adapter - NNNN CAGGTGCAGCTGAAGSASTC Universal Adapter - NNNN GAGGTCCARCTGCAACARTC Universal Adapter - NNNN CAGGTYCAGCTBCAGCARTC Universal Adapter - NNNN CAGGTYCARCTGCAGCAGTC Universal Adapter - NNNN CAGGTCCACGTGAAGCAGTC Universal Adapter - NNNN GAGGTGAASSTGGTGGAATC Universal Adapter - NNNN GAVGTGAWGYTGGTGGAGTC Universal Adapter - NNNN GAGGTGCAGSKGGTGGAGTC Universal Adapter - NNNN GAKGTGCAMCTGGTGGAGTC Universal Adapter - NNNN GAGGTGAAGCTGATGGARTC Universal Adapter - NNNN GAGGTGCARCTTGTTGAGTC Universal Adapter - NNNN GARGTRAAGCTTCTCGAGTC Universal Adapter - NNNN GAAGTGAARSTTGAGGAGTC Universal Adapter - NNNN CAGGTTACTCTRAAAGWGTSTG Universal Adapter - NNNN CAGGTCCAACTVCAGCARCC Universal Adapter - NNNN GATGTGAACTTGGAAGTGTC Universal Adapter - NNNN GAGGTGAAGGTCATCGAGTC IgG Index Reverse IgGall-IdxX-rv TruSeq Adapter Index X (RC) + Diversity region + IgG constant specific region (RC) Adapter Index X - NNNN CARKGGATRRRCHGATGGGG PE - PCR1 IgH Extension Forward Extension + VH 5' specific region IgH-extension-fw1 IgH-extension-fw2 IgH-extension-fw3 IgH-extension-fw4 IgH-extension-fw5 IgH-extension-fw6 IgH-extension-fw7 IgH-extension-fw8 IgH-extension-fw9 IgH-extension-fw10 IgH-extension-fw11 IgH-extension-fw12 IgH-extension-fw13 CCCTCCTTTAATTCCC GAKGTRMAGCTTCAGGAGTC CCCTCCTTTAATTCCC GAGGTBCAGCTBCAGCAGTC CCCTCCTTTAATTCCC CAGGTGCAGCTGAAGSASTC CCCTCCTTTAATTCCC GAGGTCCARCTGCAACARTC CCCTCCTTTAATTCCC CAGGTYCAGCTBCAGCARTC CCCTCCTTTAATTCCC CAGGTYCARCTGCAGCAGTC CCCTCCTTTAATTCCC CAGGTCCACGTGAAGCAGTC CCCTCCTTTAATTCCC GAGGTGAASSTGGTGGAATC CCCTCCTTTAATTCCC GAVGTGAWGYTGGTGGAGTC CCCTCCTTTAATTCCC GAGGTGCAGSKGGTGGAGTC CCCTCCTTTAATTCCC GAKGTGCAMCTGGTGGAGTC CCCTCCTTTAATTCCC GAGGTGAAGCTGATGGARTC CCCTCCTTTAATTCCC GAGGTGCARCTTGTTGAGTC IgH-extension-fw14 IgH-extension-fw15 IgH-extension-fw16 IgH-extension-fw17 IgH-extension-fw18 IgH-extension-fw19 IgG Extension Reverse IgGall-extension-rv CCCTCCTTTAATTCCC GARGTRAAGCTTCTCGAGTC CCCTCCTTTAATTCCC GAAGTGAARSTTGAGGAGTC CCCTCCTTTAATTCCC CAGGTTACTCTRAAAGWGTSTG CCCTCCTTTAATTCCC CAGGTCCAACTVCAGCARCC CCCTCCTTTAATTCCC GATGTGAACTTGGAAGTGTC CCCTCCTTTAATTCCC GAGGTGAAGGTCATCGAGTC Extension (RC) + IgG constant specific region (RC) GAGGAGAGAGAGAGAG CARKGGATRRRCHGATGGGG PE - PCR2 IgALL Universal Forward TruSeq Universal Adapter + Diversity region + Extension IgALL-UAd-fw Universal Adapter - NNNN CCCTCCTTTAATTCCC IgALL Index Reverse TruSeq Adapter Index (RC) + Diversity region + Extension (RC) IgALL-IdxX-rv Adapter Index X - NNNN GAGGAGAGAGAGAGAG References 1. Mazor, Y., Van Blarcom, T., Mabry, R., Iverson, B. L. & Georgiou, G. Isolation of engineered, full-length antibodies from libraries expressed in Escherichia coli. Nat Biotechnol 25, 563–565 (2007).