mec12521-sup-0002-TableS1-S3
advertisement

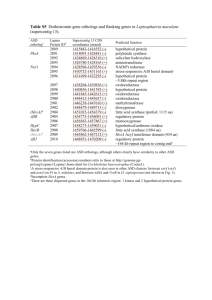
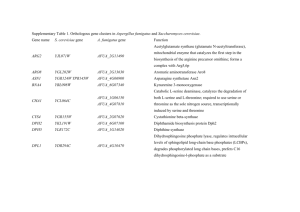
Table S1 Description of the five topo-climatic factors used in the environmental association analyses of Arabidopsis halleri. All factors are available at a resolution of 25 m and were interpolated and averaged from meteorological measurements collected over a 30-year period between 1961 and 1990 (Zimmermann & Kienast 1999). Environmental factor Abbreviation Description Unit In this study correlated with Precipitation PRECYY Yearly precipitation sum (rain and snow) 1/10 mm/yr Moisture index Slope SLP25 Slope ° Aspect* Solar radiation SRADYY Annual mean of daily global potential shortwave radiation kJ/m2/day Topographic position, topographic wetness index* Site water balance SWB Temperature TAVEYY Annual average site water balance (= precipitation – evapotranspiration) 1/10 mm/yr Moisture index, topographic wetness index* Altitude*, cloudiness, degree days, frost*, moisture index*, Mean average annual temperature 1/100 °C precipitation days, topographic position*, topographic wetness index * Negatively correlated factors. Table S2 Average genome-wide FST of pairwise population comparisons in Arabidopsis halleri, and threshold for accepting strongly differentiated SNPs. To take population structure into account, the thresholds have been adjusted by the pairwise neutral population genetic differentiation. Comparison Aha 09:11 Aha 09:19 Aha 09:21 Aha 09:31 Aha 11:19 Aha 11:21 Aha 11:31 Aha 19:21 Aha 19:31 Aha 21:31 Mean mean FST 0.042 0.034 0.035 0.026 0.041 0.048 0.044 0.037 0.035 0.036 0.038 SNP FST threshold 0.555 0.547 0.548 0.539 0.554 0.561 0.557 0.550 0.548 0.549 0.545 Table S3 The 175 identified genes covering the highly differentiated SNPs that have at least one association with one or more of the five environmental factors studied (see Table S1; Supporting information). Given are the gene IDs from TAIR10 (Lamesch et al. 2012), the number of associations with environmental factors, and the putative gene function. Gene ID AT1G04510 AT1G05540 AT1G05700 AT1G06000 AT1G06170 AT1G06960 AT1G06970 AT1G09020 AT1G09190 AT1G09970 AT1G10380 AT1G11905 AT1G15050 AT1G16460 AT1G16540 AT1G20200 AT1G20500 AT1G24095 AT1G30290 AT1G30330 AT1G48090 AT1G52900 AT1G53050 AT1G53460 AT1G53470 Precipitation (PRECYY) Slope (SLP25) SNPs associated with Radiation Site water balance (SRADYY) (SWB) 2 1 3 1 1 Temperature (TAVEYY) 8 1 2 1 1 1 1 7 6 3 2 1 1 2 1 4 5 1 1 10 1 2 4 2 2 1 1 Description Pre-mRNA-processing factor 19 hypothetical protein Leucine-rich repeat transmembrane protein kinase protein UDP-glycosyltransferase-like protein transcription factor bHLH89 RNA recognition motif-containing protein cation/H(+) antiporter 14 sucrose nonfermenting 4-like protein pentatricopeptide repeat-containing protein leucine-rich receptor-like protein kinase Putative membrane lipoprotein B-cell receptor-associated protein 31-like protein auxin-responsive protein IAA34 rhodanese homologue 2 Molybdenum cofactor sulfurase 26S proteasome regulatory subunit N3 4-coumarate--CoA ligase-like 4 Putative thiol-disulfide oxidoreductase DCC PPR repeat domain-containing protein auxin response factor 6 calcium-dependent lipid-binding-like protein Toll-Interleukin-Resistance domain-containing protein protein kinase-like protein hypothetical protein mechanosensitive channel of small conductance-like 4 AT1G53500 AT1G53520 AT1G60860 AT1G61240 AT1G64010 AT1G64070 AT1G64610 AT1G65730 AT1G68720 AT1G69720 AT1G78650 AT1G80560 AT1G80580 AT1G80660 AT1G80680 AT1G80690 AT1G80740 AT1G80750 AT2G17050 AT2G19600 1 14 AT2G19880 AT2G20960 AT2G20970 AT2G21500 AT2G21680 AT2G21770 AT2G21900 AT2G21950 AT2G21960 AT2G22070 AT2G22090 1 3 1 5 2 5 10 1 1 1 4 2 1 1 1 1 1 1 2 1 1 13 1 1 3 3 3 5 2 2 6 2 1 3 3 15 UDP-glucose 4,6-dehydratase chalcone isomerase-like protein ADP-ribosylation factor GTPase-activating protein AGD2 lysine ketoglutarate reductase trans-splicing related 1 serine protease inhibitor-like protein TIR-NBS-LRR class disease resistance protein WD40 domain-containing protein putative metal-nicotianamine transporter YSL7 tRNA-specific adenosine deaminase heme oxygenase 3 DNA-directed DNA polymerase 3-isopropylmalate dehydrogenase 2 ethylene-responsive transcription factor ERF084 H(+)-ATPase 9 suppressor of auxin resistance 3 PPPDE putative thiol peptidase family protein putative DNA (cytosine-5)-methyltransferase CMT1 60S ribosomal protein L7-1 TIR-NBS-LRR class disease resistance protein K(+) efflux antiporter 4 nucleotide-diphospho-sugar transferase domain-containing protein phospholipase-like protein (PEARLI 4) domain-containing protein putative lipid binding protein RING/U-box domain-containing protein Kelch motif-containing protein cellulose synthase A putative WRKY transcription factor 59 F-box/kelch-repeat protein SKIP6 hypothetical protein pentatricopeptide repeat-containing protein UBP1 interacting protein 1a AT2G22680 AT2G23350 AT2G23390 AT2G23460 AT2G23790 AT2G23810 AT2G24617 AT2G25410 AT2G28490 AT2G28890 AT2G29510 AT2G29760 AT2G30280 AT2G30290 AT2G30300 AT2G30590 AT2G30780 AT2G31060 AT2G31570 AT2G31620 AT2G32860 AT2G33010 AT2G33040 AT2G33150 AT2G33670 AT2G35140 AT2G35160 AT2G35800 AT2G36190 AT2G36330 AT2G36370 AT2G36910 2 31 1 1 3 3 5 1 1 3 2 2 1 14 3 4 2 1 1 2 1 8 11 2 8 1 2 6 1 2 6 1 11 2 52 1 12 1 6 C3HC4-type RING finger domain-containing protein poly(A) binding protein 4 hypothetical protein extra-large G-protein 1 hypothetical protein tetraspanin8 hypothetical protein RING-H2 finger protein ATL22 cupin domain-containing protein putative protein phosphatase 2C 23 hypothetical protein pentatricopeptide repeat-containing protein RNA-directed DNA methylation 4 vacuolar-sorting receptor 2 major facilitator protein WRKY DNA-binding protein 21 pentatricopeptide repeat-containing protein elongation factor-like protein glutathione peroxidase putative cysteine-rich repeat secretory protein 10 beta glucosidase 33 Ubiquitin-associated (UBA) protein ATP synthase subunit gamma 3-ketoacyl-CoA thiolase 2 MLO-like protein 5 DCD (Development and Cell Death) domain protein histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH5 mitochondrial substrate carrier family protein beta-fructofuranosidase hypothetical protein ubiquitin-protein ligase ABC transporter B family member 1 AT2G36950 AT2G37070 AT2G37110 AT2G38010 AT2G41460 7 16 7 AT2G41890 3 1 AT2G42330 AT2G42440 AT2G42450 AT2G42560 AT2G42570 AT2G42600 AT2G43255 AT2G43350 AT2G43370 AT2G47010 AT3G19040 AT3G19050 AT3G19220 AT3G19230 AT3G21480 AT3G22425 AT3G22430 AT3G24160 AT3G26100 AT3G26115 AT3G26120 AT3G26590 AT3G26890 AT3G27020 2 9 12 1 8 31 5 12 3 10 2 1 4 10 18 18 20 1 1 37 57 1 2 21 36 9 2 2 4 1 5 12 6 1 2 7 1 1 5 heavy-metal-associated domain-containing protein hypothetical protein PLAC8 domain-containing protein neutral ceramidase apurinic endonuclease-redox protein curculin-like mannose-binding lectin fand PAN domaincontaining protein GC-rich sequence DNA-binding factor-like protein with Tuftelin interacting domain LOB domain-containing protein 17 alpha/beta-hydrolase domain-containing protein late embryogenesis abundant domain-containing protein protein trichome birefringence-like 39 phosphoenolpyruvate carboxylase 2 hypothetical protein putative glutathione peroxidase 3 U11/U12 small nuclear ribonucleoprotein 35 kDa protein hypothetical protein transcription initiation factor TFIID subunit 1-B phragmoplast orienting kinesin 2 protein disulfide isomerase leucine-rich repeat-containing protein BRCT domain-containing DNA repair protein Imidazoleglycerol-phosphate dehydratase 1 hypothetical protein putative type 1 membrane protein regulator of chromosome condensation repeat-containing protein Pyridoxal-5'-phosphate-dependent enzyme family protein terminal EAR1-like 1 mate efflux domain-containing protein hypothetical protein putative metal-nicotianamine transporter YSL6 AT3G28315 AT3G28960 AT3G48010 AT3G49250 AT3G51390 AT3G51410 AT3G51470 AT3G51480 AT3G51490 AT3G51570 2 2 12 2 1 10 1 23 4 11 AT3G51740 AT3G51770 AT3G51850 AT3G51860 AT3G56430 AT3G56590 AT3G61750 AT3G61760 AT4G00550 AT4G00570 AT4G02010 AT4G09040 AT4G09160 AT4G10120 AT4G14220 AT4G17650 AT4G20230 AT4G21070 AT4G21450 AT4G31520 2 1 3 1 2 6 1 3 1 1 27 1 1 1 5 1 1 9 3 1 7 4 11 5 1 4 pseudo Transmembrane amino acid transporter family protein cyclic nucleotide gated channel protein defective in meristem silencing 3 putative S-acyltransferase hypothetical protein putative protein phosphatase 2C 47 glutamate receptor 3.6 monosaccharide-sensing protein 3 TIR-NBS-LRR class disease resistance protein probably inactive leucine-rich repeat receptor-like protein kinase IMK2 tetratricopeptide repeat (TPR)-containing protein calcium-dependent protein kinase 13 vacuolar cation/proton exchanger 3 hypothetical protein hydroxyproline-rich glycoprotein family protein Cytochrome b561/ferric reductase transmembrane with DOMON related domain dynamin-related protein 1B digalactosyldiacylglycerol synthase 2 malate dehydrogenase (decarboxylating) protein kinase family protein RNA recognition motif-containing protein patellin-5 sucrose-phosphate synthase E3 ubiquitin-protein ligase RHF1A Polyketide cyclase / dehydrase and lipid transport protein terpene cyclase, C1 domain-containing protein protein BREAST CANCER SUSCEPTIBILITY 1-like protein vesicle-associated membrane family protein SDA1 family protein AT5G03570 AT5G07130 AT5G11720 AT5G12210 AT5G13160 AT5G14570 AT5G14950 AT5G15340 AT5G15440 AT5G15450 AT5G37530 AT5G42800 AT5G43420 AT5G43430 AT5G43650 AT5G46270 AT5G47210 AT5G48170 AT5G48360 AT5G56680 AT5G56790 AT5G56810 AT5G62230 AT5G63000 AT5G63140 AT5G67230 AT5G67500 No annotation (various genes) 4 2 1 1 2 1 1 1 2 1 1 10 4 1 2 7 7 4 1 1 1 1 1 6 10 9 4 1 6 7 1 1 3 7 6 26 3 protein iron regulated 2 laccase 13 alpha-glucosidase RAB geranylgeranyl transferase beta subunit 1 serine/threonine-protein kinase PBS1 high affinity nitrate transporter 2.7 alpha-mannosidase II pentatricopeptide repeat-containing protein EID1-like F-box protein 1 casein lytic proteinase B3 Rossmann-fold NAD(P)-binding domain-containing protein dihydroflavonol-4-reductase RING-H2 finger protein ATL16 Electron transfer flavoprotein subunit beta transcription factor bHLH92 Disease resistance protein (TIR-NBS-LRR class) family protein hyaluronan / mRNA binding-like protein F-box protein SNE formin-like protein 9 asparaginyl-tRNA synthetase, cytoplasmic 1 protein kinase family protein putative F-box/FBD/LRR-repeat protein LRR receptor-like serine/threonine-protein kinase ERL1 Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein purple acid phosphatase 29 glycosyl transferase family 43 protein voltage dependent anion channel 2 Figure S1: Results for the three candidate genes LOS5/ABA3 (a,d,g), GPX3 (b,e,f) and ATGLR 3.6 (c,f,i). (a,b,c) represent pairwise FST values from highly differentiated sliding windows (lines) and SNPs (open circles). The bold dotted line at 0.038 indicates the mean FST across all SNPs and the dotted line at 0.47 represents the 99.9% quantile threshold for strongly differentiated sliding windows. (d,e,f) show correlations between pairwise population differences in FST, and site water balance (d, e) or solar radiation (f). rPMT represents the correlation coefficient of the partial Mantel test. (g,h,i) display linear regressions between major allele frequencies and site water balance (g, h) or solar radiation (i). References Lamesch P, Berardini TZ, Li D, et al. (2012) The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Research 40, D1202-D1210. Zimmermann NE, Kienast F (1999) Predictive mapping of alpine grasslands in Switzerland: species versus community approach. Journal of Vegetation Science 10, 469-482.