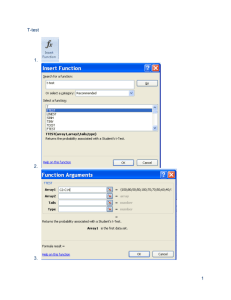
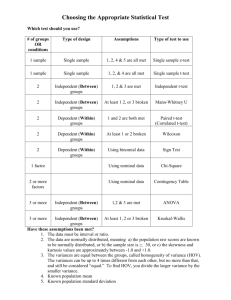
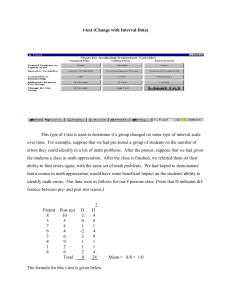
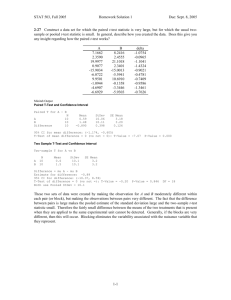
tpj12310-sup-0009-Legends
advertisement

LEGENDS to Supplemental Figures Figure S1. Transcript levels of six cytochrome c oxidase related genes. Data are the means ± SD from six independent experiments. The levels were calibrated relative to the value obtained in the GT strain under nitrogen-replete conditions, which was set at 100%. Differences between GT and GOX50 analysed using Student’s t-test were statistically significant (*P<0.05). Figure S2. Transcript levels of genes encoding PSII core proteins. Data are the mean ± SD from six independent experiments. The levels were calibrated relative to the value obtained in the GT strain under nitrogen-replete conditions, which was set at 100%. Differences between GT and GOX50 analysed using Student’s t-test were statistically significant (*P<0.05, **P<0.005). Figure S3. Transcript levels of genes encoding PSII peripheral or small accessory proteins. Data are the mean ± SD from six independent experiments. The levels were calibrated relative to the value obtained in the GT strain under nitrogen-replete conditions, which was set at 100%. Differences between GT and GOX50 analysed using Student’s t-test were statistically significant (*P<0.05, **P<0.005). Figure S4. Transcript levels of genes encoding PSII oxygen-evolving complex proteins. Data are the mean ± SD from six independent experiments. The levels were calibrated relative to the value obtained in the GT strain under nitrogen-replete conditions, which 1 was set at 100%. Differences between GT and GOX50 analyzed using Student’s t-test were statistically significant (*P<0.05, **P<0.005). Figure S5. Transcript levels of genes encoding PSI proteins. Data are the mean ± SD from independent experiments (n = 4–6). The levels were calibrated relative to the value obtained in the GT strain under nitrogen-replete conditions, which was set at 100%. Differences between GT and GOX50 analysed using Student’s t-test were statistically significant (*P<0.05, **P<0.005). Figure S6. Levels of PsbA, PsbO, and PsaD proteins. Total protein 12 g was subjected to immunoblotting. Data are the mean ± SD from three independent experiments. The expression levels were calibrated relative to the value obtained in the GT strain under nitrogen-replete conditions, which was set at 100%. Differences between GT and GOX50 analysed using Student’s t-test were statistically significant (*P<0.05). Figure S7. Levels of Rre12, Rre37, and PII proteins. (A) Purification of histidine-tagged Rre12. Histidine-tagged Rre12 (His-Rre12) purified from insoluble fraction of E. coli was subjected by SDS-PAGE and Coomassie brilliant blue staining. FT indicates flow-through fraction containing proteins unbound to resins. (B) Total protein (12 g for Rre12 and Rre37 or 6 g for PII) was subjected to immunoblotting. Data represent means ±SD of values from four independent experiments. Expression levels were calibrated relative to that in GT strain under nitrogen-replete conditions (for 2 PII) or nitrogen-depleted conditions after nitrogen depletion (for Rre12 and Rre37), which were set at 100%. Asterisks indicate significant differences between GT and GOX50 analysed using Student’s t-test (*P<0.05, **P<0.005). Figure S8. Levels of ChlH and SigA proteins. Total protein (12 g) was subjected to immunoblotting. Data represent means ±SD of values from four independent experiments. ChlI protein levels are shown as loading control. Expression levels were calibrated relative to that in GT strain under nitrogen-replete conditions, which was set at 100%. Asterisk indicates significant difference between GT and GOX50 analysed using Student’s t-test (*P<0.05). 3