IMP - University of Saskatchewan
advertisement
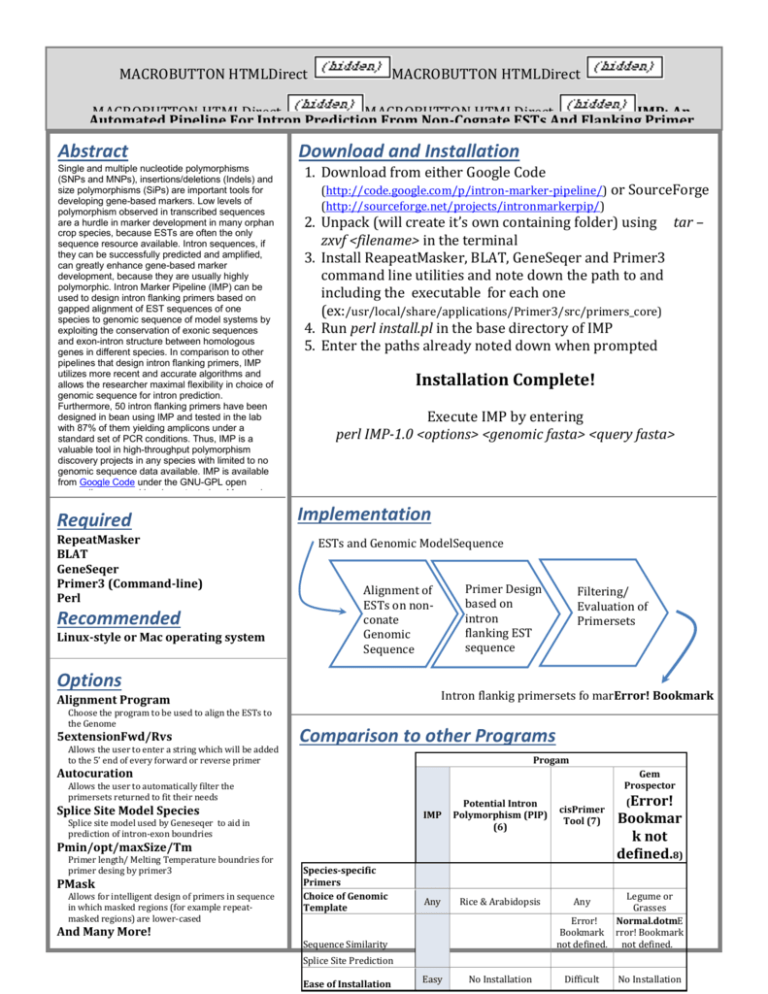
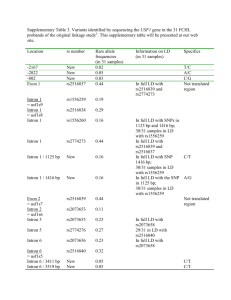
MACROBUTTON HTMLDirect MACROBUTTON HTMLDirect MACROBUTTON HTMLDirect MACROBUTTON HTMLDirect IMP: An Automated Pipeline For Intron Prediction From Non-Cognate ESTs And Flanking Primer Design To Aid In Marker Development Abstract Download Installation Lacey-Anneand Sanderson Single and multiple nucleotide polymorphisms 1. Download from either Google Code (SNPs and MNPs), insertions/deletions (Indels) and size polymorphisms (SiPs) are important tools for (http://code.google.com/p/intron-marker-pipeline/) or SourceForge developing gene-based markers. Low levels of polymorphism observed in transcribed sequences are a hurdle in marker development in many orphan crop species, because ESTs are often the only sequence resource available. Intron sequences, if they can be successfully predicted and amplified, can greatly enhance gene-based marker development, because they are usually highly polymorphic. Intron Marker Pipeline (IMP) can be used to design intron flanking primers based on gapped alignment of EST sequences of one species to genomic sequence of model systems by exploiting the conservation of exonic sequences and exon-intron structure between homologous genes in different species. In comparison to other pipelines that design intron flanking primers, IMP utilizes more recent and accurate algorithms and allows the researcher maximal flexibility in choice of genomic sequence for intron prediction. Furthermore, 50 intron flanking primers have been designed in bean using IMP and tested in the lab with 87% of them yielding amplicons under a standard set of PCR conditions. Thus, IMP is a valuable tool in high-throughput polymorphism discovery projects in any species with limited to no genomic sequence data available. IMP is available from Google Code under the GNU-GPL open source liscence and has been tested on Mac and Unix machines. Required RepeatMasker BLAT GeneSeqer Primer3 (Command-line) Perl Recommended Linux-style or Mac operating system (http://sourceforge.net/projects/intronmarkerpip/) 2. Unpack (will create it’s own containing folder) using tar – zxvf <filename> in the terminal 3. Install ReapeatMasker, BLAT, GeneSeqer and Primer3 command line utilities and note down the path to and including the executable for each one (ex:/usr/local/share/applications/Primer3/src/primers_core) 4. Run perl install.pl in the base directory of IMP 5. Enter the paths already noted down when prompted Installation Complete! Execute IMP by entering perl IMP-1.0 <options> <genomic fasta> <query fasta> Implementation ESTs and Genomic ModelSequence Options 5extensionFwd/Rvs Allows the user to enter a string which will be added to the 5’ end of every forward or reverse primer Filtering/ Evaluation of Primersets Intron flankig primersets fo marError! Bookmark not defined.ker discovery Alignment Program Choose the program to be used to align the ESTs to the Genome Primer Design based on intron flanking EST sequence Alignment of ESTs on nonconate Genomic Sequence Comparison to other Programs Progam Autocuration Gem Prospector Allows the user to automatically filter the primersets returned to fit their needs Splice Site Model Species Splice site model used by Geneseqer to aid in prediction of intron-exon boundries IMP Potential Intron Polymorphism (PIP) (6) Any Rice & Arabidopsis Easy No Installation cisPrimer Tool (7) Pmin/opt/maxSize/Tm Primer length/ Melting Temperature boundries for primer desing by primer3 PMask Allows for intelligent design of primers in sequence in which masked regions (for example repeatmasked regions) are lower-cased And Many More! Species-specific Primers Choice of Genomic Template Sequence Similarity (Error! Bookmar k not defined.8) Legume or Grasses Error! Normal.dotmE Bookmark rror! Bookmark not defined. not defined. Any Splice Site Prediction Ease of Installation Difficult No Installation