Project Description
advertisement

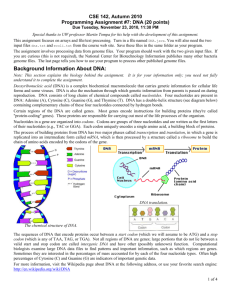
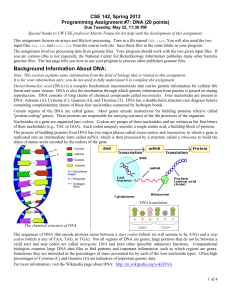
Java Project 2: DNA Analysis Students will write a program that uses arrays and files to analyze DNA sequences and determine if they represent proteins. Special thanks to Stuart Reges and Marty Stepp of UW for use of this assignment. Also thanks to Brett Wortzman of Issaquah High School for sharing the project online. I. Background Deoxyribonucleic acid (DNA) is a complex biochemical macromolecule that carries genetic information for cellular life forms and some viruses. DNA is also the mechanism through which genetic information from parents is passed on during reproduction. DNA consists of long chains of chemical compounds called nucleotides. Four nucleotides are present in DNA: Adenine (A), Cytosine (C), Guanine (G), and Thymine (T). Certain regions of the DNA are called genes. Most genes encode instructions for building proteins (they're called "protein-coding" genes). These proteins are responsible for carrying out most of the life processes of the organism. Nucleotides in a gene are organized into codons. Codons are groups of three nucleotides and are written as the first letters of their nucleotides (e.g., TAC or GGA). Each codon uniquely encodes a single amino acid, a building block of proteins. The sequences of DNA that encode proteins occur between a start codon (which we will assume to be ATG) and a stop codon (which is any of TAA, TAG, or TGA). Not all regions of DNA are genes; large portions that do not lie between a valid start and stop codon are called intergenic DNA and have other (possibly unknown) function. Computational biologists examine large DNA data files to find patterns and important information, such as which regions are genes. Sometimes they are interested in the percentages of mass accounted for by each of the four nucleotide types. Often high percentages of Cytosine (C) and Guanine (G) are indicators of important genetic data. In this assignment, you will write a program the reads named nucleotide sequences from an input file and performs analysis on the sequences. You will perform several calculations and analyses with the end goal of determining whether or not the given nucleotide sequence represents a protein. The results will be output to a file, not to the console. II. Details A. Behavior i. Program Operation Your program should being by welcoming the user and providing a brief description of the computations and analysis the program will perform. You will then prompt the user for an input file and an output file (see below for required file formats). For each nucleotide sequence in the input file, your program will compute and output the following: the number of each nucleotide (A, C, G, T) in the sequence the percentage of the sequence’s total mass accounted for by each nucleotide Page 1 of 4 the list of codons present in the sequence whether or not this sequence represents a protein (according to our rules) For our purposes, a nucleotide sequence is a protein gene if: it begins with a valid start codon (ATG), it ends with a valid stop codon (TAA, TAG, or TGA), it contains at least 5 codons total (including the start and stop codons), and Cytosine (C) and Guanine (G), combined, account for at least 30% of the sequence’s mass Note that these are not the actual constraints used by computational biologists to identify proteins; they are approximations for our assignment. The masses for each nucleotide, used for calculating the mass percentages, are as follows: Adenine (A) – 135.128 g/mol Thymine (T) – 125.107 g/mol Cytosine (C) – 111.103 g/mol Junk (-) Guanine (G) – 151.128 g/mol – 100.000 g/mol ii. Input File Format Input files for your DNA program will consist of a series pairs of lines. The first line in each pair will be a name, and the second will be a nucleotide sequence. You can assume that all input files will contain an even number of lines and will follow this format. Nucleotide sequences will consist of a series of A’s, C’s, G’s, and/or T’s (either upper- or lower-case). Nucleotide sequences can also contain dashes (‘-‘) which represent “junk” sections of the sequence. These are not nucleotides, and should be ignored when listing codons, but they do contribute mass to the sequence. iii. Output File Format For each nucleotide sequence in the input file, you should print out the name of the sequence and the nucleotides it contains. Although the input file can include either upper- or lower-case letters, your output should be entirely in upper-case. You should then output the information outlined above (nucleotide counts, mass percentages, codons, and whether or not it is a protein). For the nucleotide counts and mass percentages, you should output the values in A, C, G, T order. You must match the output format shown in the sample exactly. B. Implementation Details You must use arrays to track the various types of data you compute for each sequence. In particular, you should use arrays to keep track of the nucleotide counts, the mass percentages, and the codon list. You will use array traversals to compute each piece of data from some the others. These transformations will include: from the original nucleotide sequence to nucleotide counts from nucleotide counts to mass percentages from the original nucleotide sequence to a codon list Page 2 of 4 Your output should round the mass percentages and the total mass of the sequence to one decimal place. You can do this using either the printf method on PrintStream or the Math.round method. Look up either of these in the Java API to learn more about them. As always, your program should exhibit good functional decomposition, show good style, and be welldocumented. Make sure that your main method is a concise summary of your program’s behavior, and that you use methods (with parameters and return values) to reduce redundancy and make your program clear and readable. In addition, you should make good use of class constants in your program. Remember that class constants should be used to eliminate “magic numbers” as well as increase program readability. Take care to use class constants anywhere you are using a number that has a specific meaning. III. Input/Output File Samples A. Sample Input File cure for cancer protein ATGCCACTATGGTAG captain picard hair growth protein ATgCCAACATGgATGCCcGATAtGGATTgA bogus protein CCATt-AATgATCa-CAGTt B. Sample Output File Region Name: Nucleotides: Nuc. Counts: Total Mass%: Codons List: Is Protein?: cure for cancer protein ATGCCACTATGGTAG [4, 3, 4, 4] [27.3, 16.8, 30.6, 25.3] of 1978.8 [ATG, CCA, CTA, TGG, TAG] YES Region Name: Nucleotides: Nuc. Counts: Total Mass%: Codons List: Is Protein?: captain picard hair growth protein ATGCCAACATGGATGCCCGATATGGATTGA [9, 6, 8, 7] [30.7, 16.8, 30.5, 22.1] of 3967.5 [ATG, CCA, ACA, TGG, ATG, CCC, GAT, ATG, GAT, TGA] YES Region Name: Nucleotides: Nuc. Counts: Total Mass%: Codons List: Is Protein?: bogus protein CCATT-AATGATCA-CAGTT [6, 4, 2, 6] [32.3, 17.7, 12.1, 29.9] of 2508.1 [CCA, TTA, ATG, ATC, ACA, GTT] NO Page 3 of 4 IV. Grading Scheme/Rubric Functional Correctness (Behavior) Program greets the user and displays a description on startup Program prompts the user for input and output files Input is read from input file, not from console Output is produced in output file, not on console Output format matches specification Program prints name of sequence and list of nucleotides Program produces nucleotide counts Program produces mass percentages Program produces codon list Program determines if sequence is a protein Total 1 point 1 point 1 point 1 point 2 points 2 points 4 points 4 points 4 points 4 points 24 points Technical Correctness (Implementation) main is a concise summary of program behavior Program uses good procedural decomposition, including parameters and return values Arrays are used to track data throughout program Arrays are traversed properly to perform transformations Class constants are used where appropriate Program continues to function when constant values are changed Program uses good style and is well-documented Total 1 point 3 points Total 40 points 2 points 2 points 2 points 2 points 4 points 16 points Page 4 of 4