Managing genetic diversity within a threatened fish,
advertisement
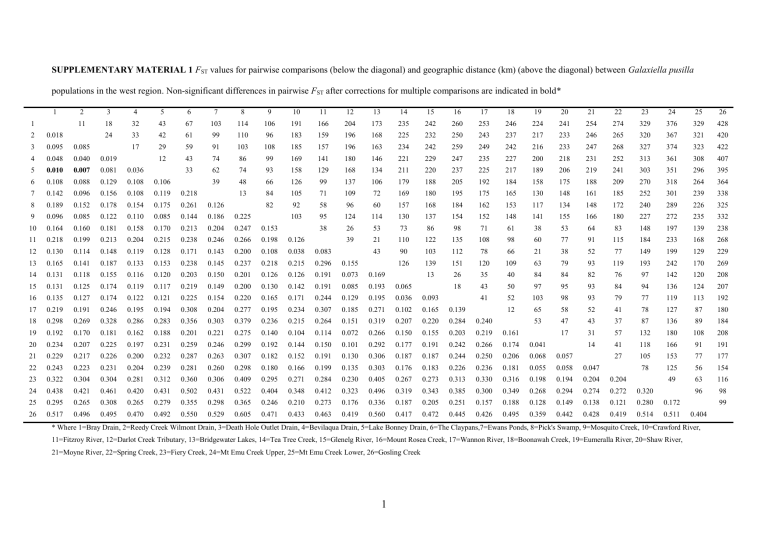
SUPPLEMENTARY MATERIAL 1 FST values for pairwise comparisons (below the diagonal) and geographic distance (km) (above the diagonal) between Galaxiella pusilla populations in the west region. Non-significant differences in pairwise FST after corrections for multiple comparisons are indicated in bold* 1 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 11 18 32 43 67 103 114 106 191 166 204 173 235 242 260 253 246 224 241 254 274 329 376 329 428 24 33 42 61 99 110 96 183 159 196 168 225 232 250 243 237 217 233 246 265 320 367 321 420 17 29 59 91 103 108 185 157 196 163 234 242 259 249 242 216 233 247 268 327 374 323 422 12 43 74 86 99 169 141 180 146 221 229 247 235 227 200 218 231 252 313 361 308 407 33 62 74 93 158 129 168 134 211 220 237 225 217 189 206 219 241 303 351 296 395 39 48 66 126 99 137 106 179 188 205 192 184 158 175 188 209 270 318 264 364 13 84 105 71 109 72 169 180 195 175 165 130 148 161 185 252 301 239 338 82 92 58 96 60 157 168 184 162 153 117 134 148 172 240 289 226 325 103 95 124 114 130 137 154 152 148 141 155 166 180 227 272 235 332 38 26 53 73 86 98 71 61 38 53 64 83 148 197 139 238 39 21 110 122 135 108 98 60 77 91 115 184 233 168 268 43 90 103 112 78 66 21 38 52 77 149 199 129 229 126 139 151 120 109 63 79 93 119 193 242 170 269 13 26 35 40 84 84 82 76 97 142 120 208 18 43 50 97 95 93 84 94 136 124 207 41 52 103 98 93 79 77 119 113 192 12 65 58 52 41 78 127 87 180 53 47 43 37 87 136 89 184 17 31 57 132 180 108 208 14 41 118 166 91 191 27 105 153 77 177 78 125 56 154 49 63 116 96 98 2 0.018 3 0.095 0.085 4 0.048 0.040 0.019 5 0.010 0.007 0.081 0.036 6 0.108 0.088 0.129 0.108 0.106 7 0.142 0.096 0.156 0.108 0.119 0.218 8 0.189 0.152 0.178 0.154 0.175 0.261 0.126 9 0.096 0.085 0.122 0.110 0.085 0.144 0.186 0.225 10 0.164 0.160 0.181 0.158 0.170 0.213 0.204 0.247 0.153 11 0.218 0.199 0.213 0.204 0.215 0.238 0.246 0.266 0.198 0.126 12 0.130 0.114 0.148 0.119 0.128 0.171 0.143 0.200 0.108 0.038 0.083 13 0.165 0.141 0.187 0.133 0.153 0.238 0.145 0.237 0.218 0.215 0.296 0.155 14 0.131 0.118 0.155 0.116 0.120 0.203 0.150 0.201 0.126 0.126 0.191 0.073 0.169 15 0.131 0.125 0.174 0.119 0.117 0.219 0.149 0.200 0.130 0.142 0.191 0.085 0.193 0.065 16 0.135 0.127 0.174 0.122 0.121 0.225 0.154 0.220 0.165 0.171 0.244 0.129 0.195 0.036 0.093 17 0.219 0.191 0.246 0.195 0.194 0.308 0.204 0.277 0.195 0.234 0.307 0.185 0.271 0.102 0.165 0.139 18 0.298 0.269 0.328 0.286 0.283 0.356 0.303 0.379 0.236 0.215 0.264 0.151 0.319 0.207 0.220 0.284 0.240 19 0.192 0.170 0.181 0.162 0.188 0.201 0.221 0.275 0.140 0.104 0.114 0.072 0.266 0.150 0.155 0.203 0.219 0.161 20 0.234 0.207 0.225 0.197 0.231 0.259 0.246 0.299 0.192 0.144 0.150 0.101 0.292 0.177 0.191 0.242 0.266 0.174 0.041 21 0.229 0.217 0.226 0.200 0.232 0.287 0.263 0.307 0.182 0.152 0.191 0.130 0.306 0.187 0.187 0.244 0.250 0.206 0.068 0.057 22 0.243 0.223 0.231 0.204 0.239 0.281 0.260 0.298 0.180 0.166 0.199 0.135 0.303 0.176 0.183 0.226 0.236 0.181 0.055 0.058 0.047 23 0.322 0.304 0.304 0.281 0.312 0.360 0.306 0.409 0.295 0.271 0.284 0.230 0.405 0.267 0.273 0.313 0.330 0.316 0.198 0.194 0.204 0.204 24 0.438 0.421 0.461 0.420 0.431 0.502 0.431 0.522 0.404 0.348 0.412 0.323 0.496 0.319 0.343 0.385 0.300 0.349 0.268 0.294 0.274 0.272 0.320 25 0.295 0.265 0.308 0.265 0.279 0.355 0.298 0.365 0.246 0.210 0.273 0.176 0.336 0.187 0.205 0.251 0.157 0.188 0.128 0.149 0.138 0.121 0.280 0.172 26 0.517 0.496 0.495 0.470 0.492 0.550 0.529 0.605 0.471 0.433 0.463 0.419 0.560 0.417 0.472 0.445 0.426 0.495 0.359 0.442 0.428 0.419 0.514 0.511 99 0.404 * Where 1=Bray Drain, 2=Reedy Creek Wilmont Drain, 3=Death Hole Outlet Drain, 4=Bevilaqua Drain, 5=Lake Bonney Drain, 6=The Claypans,7=Ewans Ponds, 8=Pick's Swamp, 9=Mosquito Creek, 10=Crawford River, 11=Fitzroy River, 12=Darlot Creek Tributary, 13=Bridgewater Lakes, 14=Tea Tree Creek, 15=Glenelg River, 16=Mount Rosea Creek, 17=Wannon River, 18=Boonawah Creek, 19=Eumeralla River, 20=Shaw River, 21=Moyne River, 22=Spring Creek, 23=Fiery Creek, 24=Mt Emu Creek Upper, 25=Mt Emu Creek Lower, 26=Gosling Creek 1 SUPPLEMENTARY MATERIAL 2 FST values for pairwise comparisons (below the diagonal) and geographic distance (km) (above the diagonal) between Galaxiella pusilla populations in the east region. Non-significant differences in pairwise FST after corrections for multiple comparisons are indicated in bold* 27 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 12 15 17 26 34 26 17 56 57 73 38 114 128 172 179 201 207 227 264 149 356 379 429 266 12 16 27 34 24 6 47 47 63 26 104 118 161 169 191 197 218 255 137 344 368 417 260 5 15 22 12 18 57 57 71 31 113 126 168 177 199 206 228 265 140 348 368 419 251 11 18 8 22 61 61 75 35 117 130 172 182 204 211 233 270 143 351 371 421 249 7 7 33 72 71 84 44 127 139 180 191 213 220 242 279 148 357 373 425 241 11 40 77 77 89 48 131 143 184 195 217 224 247 284 149 358 372 425 235 29 66 65 78 37 120 133 173 184 206 213 236 273 141 350 367 418 240 40 41 57 22 98 111 155 163 185 191 212 249 133 339 364 413 263 4 21 31 59 72 117 123 146 151 172 209 107 306 339 385 277 18 29 57 71 115 122 144 150 171 209 103 303 335 382 274 41 42 55 98 106 129 135 158 195 87 286 319 364 271 83 95 137 147 169 176 199 236 111 318 342 391 251 14 60 65 87 93 117 154 74 252 296 337 292 46 52 74 81 106 143 70 239 285 325 296 27 38 54 83 116 65 193 245 282 302 23 31 59 94 91 202 262 295 328 20 48 79 102 182 249 278 338 29 63 119 194 265 292 356 37 148 202 281 303 385 181 196 285 300 416 209 232 280 237 103 103 367 61 314 28 0.182 29 0.301 0.175 30 0.308 0.171 0.278 31 0.151 0.158 0.164 0.235 32 0.335 0.176 0.132 0.253 0.154 33 0.478 0.308 0.232 0.418 0.276 0.107 34 0.505 0.484 0.438 0.506 0.305 0.432 0.573 35 0.486 0.504 0.433 0.554 0.277 0.441 0.581 0.212 36 0.442 0.427 0.373 0.471 0.237 0.374 0.507 0.051 0.101 37 0.600 0.602 0.571 0.644 0.446 0.569 0.684 0.234 0.310 0.198 38 0.437 0.451 0.392 0.488 0.244 0.396 0.530 0.070 0.079 0.032 0.201 39 0.547 0.532 0.512 0.587 0.398 0.511 0.604 0.536 0.494 0.461 0.592 0.449 40 0.509 0.505 0.483 0.549 0.354 0.487 0.597 0.503 0.461 0.431 0.568 0.412 0.227 41 0.566 0.603 0.620 0.648 0.518 0.626 0.722 0.659 0.644 0.603 0.679 0.597 0.643 0.617 42 0.461 0.505 0.492 0.558 0.370 0.523 0.623 0.477 0.446 0.395 0.530 0.404 0.445 0.415 0.569 43 0.387 0.408 0.381 0.442 0.275 0.398 0.504 0.405 0.356 0.331 0.444 0.327 0.307 0.249 0.491 0.169 44 0.470 0.469 0.433 0.495 0.300 0.433 0.526 0.425 0.381 0.354 0.498 0.338 0.183 0.097 0.590 0.332 0.161 45 0.559 0.569 0.502 0.613 0.400 0.538 0.652 0.524 0.480 0.442 0.596 0.430 0.398 0.325 0.683 0.337 0.178 0.177 46 0.537 0.548 0.521 0.596 0.362 0.523 0.654 0.484 0.441 0.398 0.572 0.382 0.332 0.258 0.667 0.353 0.224 0.142 0.264 47 0.510 0.512 0.517 0.583 0.409 0.559 0.655 0.550 0.567 0.495 0.615 0.481 0.556 0.522 0.604 0.562 0.461 0.505 0.630 0.611 48 0.648 0.646 0.623 0.723 0.512 0.667 0.775 0.711 0.701 0.644 0.790 0.629 0.688 0.625 0.794 0.683 0.592 0.599 0.746 0.755 0.627 49 0.437 0.390 0.421 0.416 0.326 0.436 0.549 0.461 0.486 0.416 0.538 0.434 0.551 0.508 0.591 0.519 0.424 0.455 0.569 0.539 0.535 0.585 50 0.426 0.430 0.438 0.496 0.316 0.483 0.590 0.519 0.515 0.456 0.601 0.457 0.567 0.524 0.577 0.467 0.413 0.486 0.581 0.547 0.521 0.675 0.409 0.277 0.379 0.446 0.501 0.302 0.477 0.637 0.603 0.584 0.526 0.686 0.518 0.622 0.588 0.613 0.505 0.429 0.538 0.672 0.646 0.568 0.811 0.473 51 374 0.463 * Where 27=Tributary of Dandenong Creek, 28=Golf Links Road Drain, 29=Eastern Contour Drain Tributary, 30=Boggy Creek, 31=Balcombe Creek, 32=Tuerong Creek, 33=Blackscamp Wetland, 34=Cardinia Creek Tributary, 35=Dingo Creek Tributary, 36=Cannibal Creek, 37=King Parrot Creek, 38=Yallock Creek, 39=Moe Contour Drain, 40=Morwell River, 41=Deep Creek, 42=Merriman Creek, 43=Monkey Creek, 44=Flooding Creek, 45=Perry River, 46=Cobblers Creek, 47=Darby River , 48=Flinders Island, 49=Big Waterhouse Lake, 50=Icena Creek, 51=Harcus River 2 SUPPLEMENTARY MATERIAL 3 Two-dimensional plot showing the relationships among individuals of Galaxiella pusilla based on a multivariate factorial correspondence analysis of 10 loci from (a) the west region and (b) the east region. The first two factors are shown, with the percentage of variance explained in parenthesis. See Tables 1 & 2 for translation of site codes. (a) West Region 3000 2500 Factor 2 (10.23%) 2000 1500 1000 500 0 -500 -1000 -1500 -1500 -1000 -500 0 500 1000 1500 Factor 1 (16.42%) BRD RCW DEH BEV LBD CLP EWP PIS MOS CRR FIT DLT BRL TTC GLR MRC WAR BOO EUR SHR MOY SPR FIC MEU MEL GOC (b) East Region 2000 1500 Factor 2 (12.93%) 1000 500 0 -500 -1000 -4000 -3000 -2000 -1000 Factor 1 (15.40%) 3 0 1000 TDN GLD ECD BOC BAC TUC BCW CAR DIN CAN KPC YAC MOE MOR DEE MER MON FLC PER COB DAR WIN BWL ICC HAR SUPPLEMENTARY MARTERIAL 4 Bayesian inference phylogenetic trees for Galaxiella pusilla haplotypes generated from (a) 16S rDNA (16S), (b) cytochrome c oxidase subunit I (COI) and (c) Cytochrome b (CytB) fragments of 482bp, 595bp, and 1083bp in size, respectively. The black-stripe minnow, Galaxiella nigrostriata, and common galaxias, Galaxias maculatus, were used as outgroups. See Tables 1 & 2 for translation of site codes. Comments on tree construction For the construction of Bayesian inference phylogenetic trees, four Metropolis-coupled Monte Carlo Markov Chains (MCMC) were run for 5,000,000 generations, and trees were sampled every 100 generations. A temperature parameter of either 0.1 or 0.2 was applied to ensure that heated chains were all successfully contributing possible trees. Convergence of runs was considered complete when the average standard deviation of split frequencies between duplicate runs was less than 0.01. Stationarity of the chains was assessed by plotting log likelihood values against generation time and observing the point where a stable mean equilibrium value was reached. Burn in values were conservatively set to discard the first 1,250 trees (to remove trees outside the stable mean equilibrium) prior to calculation of a consensus topology and estimates of posterior probabilities for 50% majority rule consensus trees. Bayesian posterior probabilities greater than 0.95 were considered significant support for a clade (Hillis and Bull 1993; Schultz et al. 2009). References Hillis, D.M., Bull, J.J. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology, 42, 182–192. Schultz MB, Smith SA, Horwitz P, Richardson AMM, Crandall KA, Austin CM (2009) Evolution underground: A molecular phylogenetic investigation of Australian burrowing freshwater crayfish (Decapoda: Parastacidae) with particular focus on Engaeus Erichson. Molecular Phylogenetics and Evolution, 50, 580–598 4 a) 16S 16S Hap6 (SHR) 16S Hap2 (FIC) 16S Hap13 (All other west region sequences) 100 16S Hap4 (BEV4) 16S Hap5 (BEV,DEH) 99 West 16S Hap1 (FIT) 92 99 16S Hap9 (CRR,DLT) 91 16S Hap8 (DLT2) 99 16S Hap10 (DLT4) 16S Hap7 (BRD6) 87 16S Hap15 (BWL,DAR,ECD1,HAR,ICC) 16S Hap11 (WIN) 100 East 16S Hap14 (PER) 16S Hap13 (All other east region sequences) 16S Hap12 (FLC2) Galaxiella nigrostriata Galaxias maculatus 0.005 5 b) COI CO1 Hap1 (DLT7) CO1 Hap35 (DLT4,9) CO1 Hap36 (DLT1) CO1 Hap25 (BOO,EUM,MOY,SHR,SPR) CO1 Hap26 (EUM1,8) 79 91 CO1 Hap21 (CRR6,9,10) CO1 Hap20 (CRR) CO1 Hap11 (DLT) 60 CO1 Hap9 (FIT2,3,6) Fitzroy CO1 Hap10 (FIT) 98 CO1 Hap13 (GOC) CO1 Hap2 (TTC05) CO1 Hap3 (BRD4,8) 99 84 CO1 Hap5 (LBD3,10,RCW3) CO1 Hap6 (BRD,CLP,LBD,RCW) CO1 Hap7 (BEV7) 50 Bevilaqua Death Hole Fiery CO1 Hap27 (BEV,DEH) 99 CO1 Hap32 (BEV1) CO1 Hap4 (FIC4) CO1 Hap8 (FIC) 99 Bray, Lake Bonney, Claypans, Reedy Ck West CO1 Hap12 (TTC1,8) CO1 Hap14 (MRC5,7) CO1 Hap22 (GLR,MRC,TTC7) 60 CO1 Hap23 (MOS9) 82 CO1 Hap24 (MOS) 98 CO1 Hap16 (WAR) Wannon CO1 Hap15 (WAR5) 100 CO1 Hap17 (GLR3,7,8,MRC2,TTC) CO1 Hap18 (BRL4) CO1 Hap19 (BRL) CO1 Hap28 (MEU) CO1 Hap29 (MEL) 86 CO1 Hap30 (MEL9) 78 CO1 Hap31 (MEL8) Port Phillip Western Port CO1 Hap33 (EWP,PIS) 86 CO1 Hap34 (PIS9) CO1 Hap37 (YAC4) 95 CO1 Hap38 (BAC,BCW,BOC,CAN,CAR,DIN,ECD,GLD,KPC,TDN,TUC,YAC) CO1 Hap39 (KPC4) CO1 Hap40 (MOR1,4,11) CO1 Hap48 (FLC8,9) 74 CO1 Hap50 (COB,FLC,MOE,MOR) CO1 Hap41 (MON1) CO1 Hap42 (WIN) 100 53 CO1 Hap44 (BWL,ICC) 98 CO1 Hap47 (BWL5) CO1 Hap45 (HAR) 97 CO1 Hap46 (DAR) CO1 Hap43 (PER) CO1 Hap49 (FLC2) Galaxiella nigrostriata Galaxias maculatus 0.02 6 East CO1 Hap51 (DEE,MER,MON) 71 Wilsons Promontory Tasmania c) CytB CYTB Hap72 (FIC9) CYTB Hap70 (FIC5) CYTB Hap69 (FIC1) CYTB Hap71 (FIC) CYTB Hap68 (FIC7) CYTB Hap82 (GOC) CYTB Hap77 (MEL10,11) 100 CYTB Hap76 (MEL) 89 Mt Emu CYTB Hap75 (MEU) CYTB Hap58 (DEH8) CYTB Hap43 (BEV,DEH) CYTB Hap49 (RCW2) CYTB Hap47 (CLP) CYTB Hap46 (LBD4) CYTB Hap45 (BRD10,LBD1,RCW6,7,8) 100 100 South Australia CYTB Hap44 (BEV7) CYTB Hap81 (BRD2,5,9) 73 99 CYTB Hap80 (RCW5) CYTB Hap79 (BRD,LBD,RCW1,3,10,11) CYTB Hap48 (BRD7) 93 CYTB Hap42 (BRD3,LBD5,7,RCW9) CYTB Hap41 (DEH5) CYTB Hap40 (DEH6) West CYTB Hap39 (WAR2) 100 Wannon CYTB Hap38 (WAR) CYTB Hap33 (BRL) CYTB Hap37 (EWP1) 9694 Bridgewater Ewans, Picks CYTB Hap36 (EWP10) CYTB Hap35 (EWP2,9) 100 87 CYTB Hap34 (PIS) CYTB Hap29 (EWP) CYTB Hap56 (BOO,MOY,SPR) 93 CYTB Hap55 (EUM) CYTB Hap54 (EUM39) 98 91 Eumeralla CYTB Hap53 (EUM35) CYTB Hap32 (MOY31) CYTB Hap31 (SHR) CYTB Hap30 (MOY32,36,40,SHR31) 98 CYTB Hap66 (FIT4) 90 100 CYTB Hap63 (FIT) Fitzroy CYTB Hap74 (FIT1) 99 CYTB Hap59 (FIT7) CYTB Hap64 (DLT31,37,39) CYTB Hap65 (CRR36,39,40) 73 95 CYTB Hap67 (CRR32) Crawford CYTB Hap62 (CRR) 100 CYTB Hap78 (DLT33) 91 86 CYTB Hap61 (DLT38) CYTB Hap60 (DLT35,36,40) 96 CYTB Hap73 (DLT34) CYTB Hap28 (DLT32) 92 CYTB Hap51 (MOS) Grampians Mosquito CYTB Hap57 (GLR35) 100 CYTB Hap52 (MRC7) CYTB Hap50 (GLR,MRC,TTC) 71 CYTB Hap27 (GLR31,32,40,MRC1,8,10,TTC7) CYTB Hap20 (HAR) CYTB Hap26 (ICC) 100 CYTB Hap25 (WIN5) 100 Tasmania Wilsons Promontory CYTB Hap24 (WIN8,9,10) CYTB Hap23 (WIN3) 88 100 CYTB Hap22 (WIN) CYTB Hap21 (BWL4,6) CYTB Hap19 (BWL) 99 CYTB Hap18 (BWL2) CYTB Hap17 (BWL8) CYTB Hap16 (DAR39) East CYTB Hap1 (DAR) 97 91 CYTB Hap15 (DEE) Deep CYTB Hap14 (DEE2,3,6,7) CYTB Hap13 (MER) 100 57 CYTB Hap12 (FLC1,3) CYTB Hap11 (COB36) CYTB Hap10 (COB,FLC,MOE,MON,MOR) CYTB Hap9 (MOE31,35,41) CYTB Hap7 (PER) CYTB Hap8 (BOC3,6,11) CYTB Hap6 (TDN) Port Phillip Westernport CYTB Hap5 (CAN11) 98 CYTB Hap4 (CAR39) CYTB Hap3 (GLD9) CYTB Hap2 (BAC,BCW,BOC,CAN,CAR,DIN,ECD,GLD,KPC,TDN3,6,10,TUC,YAC) Galaxiella nigrostriata Galaxias maculatus 0.02 7 SUPPLEMENTARY MATERIAL 5 Analysis of demographic history of Galaxiella pusilla populations, where populations are grouped according to east and west regions. Significant values after correction for multiple comparisons are in bold font. N Tajima’s D Pvalue Mismatch Growth-Decline Raggedness index r P-value Fu & Li’s F* P-value Fu & Li’s D* P-value Fu’s Fs P-value 16S 10 -1.0143 0.1850 0.1027 0.2896 -1.1288 0.1900 -1.0008 0.1940 -9.412 <0.001 COI 36 -0.8711 0.1780 0.0073 0.0080 -0.4273 0.3250 -0.0964 0.4400 -37.057 <0.001 CytB 56 -1.1396 0.1140 =0,1000 2ut=2.4444 =2.44492,1000 2ut=5.9662 =4.37590,1000 2ut=10.1676 0.0034 <0.0001 -1.9822 0.0370 -1.9751 0.0330 -34.117 <0.001 16S 5 -0.4102 0.4440 0.2400 0.4625 -0.4175 0.4340 -0.4102 0.4310 -3.304 0.035 COI 15 -0.8235 0.2180 0.1005 0.6356 -1.1297 0.1610 -1.0428 0.1450 -12.153 <0.001 CytB 27 0.1560 0.6270 =0,1000 2ut=1.8 =1.10567,1000 2ut=3.8086 =4.81871,1000 2ut=5.9904 0.0116 0.0440 0.0295 0.5280 -0.0365 0.4280 -16.201 <0.001 WEST EAST 8 SUPPLEMENTARY MATERIAL 6 Two-dimensional plot comparing genetic diversity (mean number of alleles and heterozygosity) to genetic uniqueness (mean Cytochrome b Kimura-2-parameter distance) for each Galaxiella pusilla population within the west region (a) and (b) and the east region (c) and (d) . See Tables 1& 2 for translation of site codes (a) (b) 0,018 R² = 0,0981 0,018 0,017 0,016 0,016 0,015 0,015 Mean CytB K2P Mean CytB K2P 0,017 0,014 0,013 0,012 R² = 0,1513 0,014 0,013 0,012 0,011 0,011 0,010 0,010 0 2 4 6 8 Mean No. Alleles 0,1 10 (c) 0,3 0,5 Heterozygosity (HE) 0,7 (d) 0,013 0,013 R² = 0,1845 0,012 R² = 0,2569 0,012 0,011 Mean CytB K2P Mean CytB K2P 0,011 0,010 0,009 0,010 0,009 0,008 0,008 0,007 0,007 0,006 0,006 1 2 3 4 Mean No. Alleles 5 0 9 0,1 0,2 0,3 0,4 Heterozygosity (HE) 0,5