mec12428-sup-0001-FigS1-TableS1-3
advertisement

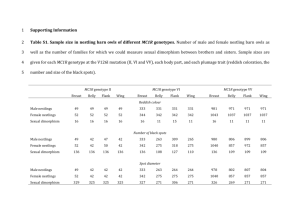
Supporting information S1 Substitution models Best-fit models of nucleotide substitution were determined for MC1R and neutral loci in jModeltest version 0.1.1 (Posada 2008), using likelihood calculations with optimized ML and the Akaike information criterion (AIC). Calculations included models with equal or unequal base frequencies (+F) and rate variation among sites with a number of rate categories (+G) only, because models with a proportion of invariable sites (+I) are not included in Arlequin version 3.5.1.2 (Excoffier & Lischer 2010), that was used for calculating nucleotide diversities. Either best-fit or the nearest less-complex models were chosen (MC1R, Lamin and RP40: TrN+G, α = 0.01; MPP: TrN+G, α = 0.02 and mtDNA: JC). S2 Probability of non-synonymous substitutions in MC1R being concentrated near root of network in arctic skuas 7 6 5 4 NonSynonymous 3 Synonymous 2 1 0 0 1 2 3 Fig. S1 Frequency distribution of distance (x-axis) of non-synonymous and synonymous substitutions from the ancestral haplotype in the arctic skua MC1R network (Figure 2). Total number of substitutions = 17. To calculate the exact probability of four mutations occupying the bins at 0 steps (2 mutations) and 1 step (2 mutations) from the root, we considered the potential paths in which 4 sequential randomly placed mutations would lead to the desired distribution. There are 6 possible paths (unoccupied bins are represented by ” –“ and dots represent mutations occupying bins): 1. () → () → ( ) → ( ) 2. () → ( ) → ( ) → ( ) 3. () → ( ) → ( ) → ( ) 4. ( ) → ( ) → ( ) → ( ) 5. ( ) → ( ) → ( ) → ( ) 6. ( ) → () → ( ) → ( ) Each of these paths has the same probability given by (1 x 2 x 7 x 8)/(17 x 16 x 15 x 14) = 1/510, so the total probability is 6 x 1/510 = 1/85 = 0.0118 (3 sig figs). This was verified by calculating the total number of ways of placing 4 mutations in bins 0 and 1 divided by the total number of ways 4 mutations could be distributed among the 4 bins. Table S1: Sampling details a Location Coordinates Collection year Number of DNA samples Material 1 Colville Rivera 70.00 N, 151.00 W 1998 17 2 Åland 60.13 N, 20.15 E 1999/2002 26 3 Slettnes 71.05 N, 28.13 E 1997/98 23 4 Bear Island 74.26 N, 19.10 E 2000 22 5 Spitsbergen 78.56 N, 11.53 E 2000 18 Tissue in DSMObuffer Blood in SET-buffer Blood in SET-buffer Blood in SET-buffer Blood in SET-buffer Number of individuals with phenotypic data 17 38 322 104 60 Tissue samples and plumage colour morph data obtained from Alaska Frozen Tissue Collection, University of Alaska Museum, Fairbanks, USA Table S2 MC1R and intron haplotypes and their frequencies in five arctic skua populations. Variable sites are shown. Dots indicate identity with the first haplotype presented. Non-synonymous substitutions for MC1R are shown in bold. SP is arctic skua and SL is longtailed skua. Nucleotide position MC1R Alleles (Genbank accessions KF268961KF268987) SP1 pale SP2 pale SP3 pale SP4 pale SP5 pale SP6 pale SP7 pale SP10 pale SP12 pale SP13 pale SP14 pale SP15 pale SP16 pale SP8 melanic SP9 melanic SP11 melanic SL1 000000000000000000000000000000000001 001111222222333333333455666667888990 890046055669012366789913366890299570 331402404234889039247207634268817742 CGCGTCGCCGGAACGCCCCCAGCGTCACCCCGCGGG .................................A.. .............................T...... ....................G............... ....................G.T............. ....................GA.............. .................T.....A....T....... ..........C......................... ....C............................... ....C........................T...... ..G................................. T................................... T.........C......................... ............G.A...................A. ............G.A..................AA. .......T....G.A...................A. (1) (2) (3) Alaska Åland Slettnes 7 0 0 6 0 1 0 1 0 0 0 0 0 15 0 4 8 0 0 6 0 0 0 2 0 0 0 0 0 32 1 3 12 4 0 5 1 0 0 1 0 0 2 1 20 0 0 (4) Bear Island (5) Spitzbergen total frequency 25 0 0 10 0 0 1 5 0 0 2 1 0 0 0 0 24 0 1 7 0 0 0 3 0 1 0 0 0 0 0 0 76 4 1 34 1 1 1 11 1 1 2 3 1 67 1 7 3 SL2 SL3 SL4 SL5 SL6 SL7 SL8 SL9 SL10 SL11 Lamin Alleles (Genbank accessions KF268999KF269006) SP1 SP2 SP3 SP4 SP5 SP6 SP7 SP8 MPP Alleles ........G..GG...........C.G...T....A ........G..GG...........C.G......... ........G..GG....T......C.G.T.TA...A ........G..GG..T........CTGT........ ........G..GGT..........C.G...T....A ........GA.GG...........C.G...T....A ......A.GA.GG...........C.G......... ......A.GA.GG.....T.....C.G......... .....T..G..GG...T.TT....CTG......... ...A....G..GG...........CTG...T..... .A......G..GG...........CTG.....T... 3 2 2 3 2 1 1 1 1 1 Nucleotide position (1) (2) (3) 0000111 0589019 8087278 Alaska Åland CCACGGC ......T ....AAT ..G.... ..G...T ..GG... .T....T T...... 16 10 2 3 0 1 2 0 11 24 0 14 3 0 0 0 Nucleotide position Gamvik (4) Bear Island (5) Spitzbergen total frequency 22 17 0 4 1 2 0 0 28 5 0 8 0 1 0 2 16 17 0 2 0 0 0 1 93 73 2 31 4 4 2 3 (Genbank accessions KF269007KF269026) SP1 SP2 SP3 SP4 SP5 SP6 SP7 SP8 SP9 SP10 SP11 SP12 SP13 SP14 SP15 SP16 SP17 SP18 SP19 SP20 * 00001112223 35991894792 15793998017 (1) (2) (3) Alaska Åland AGACGAGGGCC ........A.. ....A..A... ....A..A..T ..-........ G.......... G........T. G.......A.. G.......AT. G......A..T G.....A.... G...A...... G...A..A... G...A..A..T G...AG..... G..T....... G.-........ GA......... GA......A.. GA..A...A.. 11 0 1 1 0 14 0 3 0 0 0 0 0 3 0 1 0 0 0 0 33 0 0 0 1 7 0 2 0 2 1 1 0 1 2 0 2 0 0 0 * deletion of 8 bp RP40 Nucleotide position Gamvik (4) Bear Island (5) Spitzbergen total frequency 12 1 0 0 0 6 0 0 6 0 0 1 0 6 0 1 0 1 11 1 16 0 1 0 0 5 0 0 1 0 0 0 3 14 0 0 3 0 1 0 5 0 0 0 0 14 1 1 1 0 0 0 3 9 0 0 2 0 0 0 77 1 2 1 1 46 1 6 8 2 1 2 6 33 2 2 7 1 12 1 Alleles (Genbank accessions KF268988KF268998) SP1 SP2 SP3 SP4 SP5 SP6 SP7 SP8 SP9 SP10 SP11 (1) (2) (3) Gamvik (4) Bear Island (5) Spitzbergen 01111223 41126342 64778159 Alaska Åland total frequency ACGGGACC ......T. ....C.T. ...-..T. ...-.GT. .T...... .T....T. .T....TT .TC...T. .TC...TT G.....T. 8 11 2 2 1 1 6 3 0 0 0 13 29 8 0 0 0 0 0 0 2 0 13 17 0 0 0 1 12 1 0 2 0 4 24 0 0 0 0 8 6 1 0 1 12 14 0 0 0 1 6 3 0 0 0 50 95 10 2 1 3 32 13 1 4 1 Table S3 Tajima’s D and Fu’s FS for five arctic skua populations, using standardized sample sizes (N = 17 individuals per population). Alaska Åland Slettnes Bear I. Spitz Overall D MC1R Lamin MPP RP40 mtDNA 0.805 -0.667 -0.048 -0.253 -0.973 0.625 1.897 -1.274 -0.835 -0.049 0.337 0.405 0.665 0.404 0.243 -1.179 -0.882 0.375 -0.176 -1.047 -0.769 -0.313 0.180 1.024 -0.991 FS MC1R -0.882 -1.162 -1.290 -2.524 -6.952** 1.168 0.419 -5.859** 0.081 -0.357 0.032 -0.881 0.438 -1.101 -2.524 -1.458 -1.722 -0.721 -0.748 -2.461 -1.538 -2.583 -0.573 -1.938 -0.548 -6.186 -0.331 -2.775 -1.009 -25.320** Lamin MPP RP40 mtDNA ** p < 0.01 -0.735 -0.592 0.028 -0.241 -0.851




![WIA poetry exercise[1]](http://s3.studylib.net/store/data/006641954_1-9e009b6b11d30a476e3c12c4ba5f8bb1-300x300.png)