nonparametric
advertisement

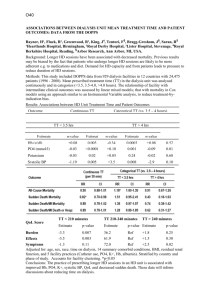
Correlation and non-parametric tests # Download set of gene expression data ge <- read.table(“http://www.ebi.ac.uk/goldmansrv/PredocCourse2009/gene_expression.txt”, header=TRUE) ge[1:5,] ProbeName 1 Exp1 Affymetrix:CompositeSequence:ATH1-121501:AFFX-Athal-5SrRNA_at 12.4 2 Affymetrix:CompositeSequence:ATH1-121501:AFFX-Athal-Actin_3_f_at 1297.0 3 Affymetrix:CompositeSequence:ATH1-121501:AFFX-Athal-Actin_5_r_at 4 73.9 Affymetrix:CompositeSequence:ATH1-121501:AFFX-Athal-Actin_M_at 943.6 5 Affymetrix:CompositeSequence:ATH1-121501:AFFX-Athal-GAPDH_3_s_at 1301.4 1 Exp2 Exp3 Exp4 Exp5 3.6 0.9 2.1 3.4 Exp6 12.0 2 1354.8 1401.5 1198.6 1017.4 1322.2 3 83.4 87.4 156.4 150.3 69.0 4 938.9 904.8 1133.4 958.2 940.1 5 1089.4 1153.5 1173.5 1157.8 1337.5 summary(ge$Exp1) Min. 1st Qu. Median 0.0 9.0 38.6 Mean 3rd Qu. 135.2 Max. 119.5 4759.0 # Prepare log-scale data (must add small constant because # of zero values lge1 <- log(ge$Exp1 + 0.001) lge2 <- log(ge$Exp2 + 0.001) # Basic plot. See oddities due to rounding at left—hand # side plot( lge1 , lge2 , pch=’.’) # Test correlation. Pearson is default method cor.test(lge1,lge2) Pearson's product-moment correlation data: lge1 and lge2 t = 573.0363, df = 22808, p-value < 2.2e-16 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: 0.9661278 0.9678138 sample estimates: cor 0.9669814 # Different scales cor.test(lge1,ge$Exp2) Pearson's product-moment correlation data: lge1 and ge$Exp2 t = 98.9823, df = 22807, p-value < 2.2e-16 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: 0.5390315 0.5571884 sample estimates: cor 0.5481746 # Contrast to Spearman cor.test(lge1,ge$Exp2,method="spearman") Spearman's rank correlation rho data: lge1 and ge$Exp2 S = 44539905431, p-value < 2.2e-16 alternative hypothesis: true rho is not equal to 0 sample estimates: rho 0.9774793 Warning message: In cor.test.default(lge1, ge$Exp2, method = "spearman") : Cannot compute exact p-values with ties # We get warning because calculations for exact # distribution of Spearman cannot cope with tied data. # For this many samples it uses an approximation in # any case. # Remember that Spearman is just Pearson with ranks cor.test(rank(lge1),rank(ge$Exp2)) Pearson's product-moment correlation data: rank(lge1) and rank(ge$Exp2) t = 699.5114, df = 22807, p-value < 2.2e-16 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: 0.976894 0.978050 sample estimates: cor 0.9774793 # Could also investigate Kendall. Takes longer to # calculate. Look at over all distribution of gene expression and see if there are any discernable differences. # Plot an “empirical cumulative density graph” # i.e. rank against value plot( ecdf(lge1) , pch=’.’) # Add second set of expression data lines( ecdf(lge2) , pch=’.’) # Can see that the two graphs diverge for low expression ks.test(lge1,lge2) Two-sample Kolmogorov-Smirnov test data: lge1 and lge2 D = 0.0207, p-value = 0.0001146 alternative hypothesis: two-sided Warning message: In ks.test(lge1, lge2) : cannot compute correct p-values with ties # Check different alternatives. Difference is one-sided ks.test(lge1,lge2,alternative=”less”)$p.value [1] 0.8757951 ks.test(lge1,lge2,alternative="greater")$p.value [1] 5.729068e-05 # One experiment has consistently higher expression # Look at differences between two experiments # Genes form natural pairing, so use Wilcoxon Signed Rank # conf.int=TRUE means report confidence intervals # Test reports the pseudo median – the median of the # differences between the two sets of expression data wilcox.test( lge1 , lge2 , paired=TRUE, conf.int=TRUE) Wilcoxon signed rank test with continuity correction data: lge1 and lge2 V = 108321954, p-value < 2.2e-16 alternative hypothesis: true location shift is not equal to 0 95 percent confidence interval: -0.05212866 -0.04213704 sample estimates: (pseudo)median -0.04713760 # If we ran same test using non-logged data we’d get a # slightly different result since Signed Rank test ranks # the differences not the actual values. # Ignore genes and just look for changes in expression # between experiments (Wilcoxon Rank Sum). # P-value significant but much less. Knowing the # gene-pairs provides much more info. wilcox.test(lge1,lge2,paired=FALSE) Wilcoxon rank sum test with continuity correction data: lge1 and lge2 W = 256220403, p-value = 0.005493 alternative hypothesis: true location shift is not equal to 0 # Actually have 6 experiments. Instead of Wilcox Rank Sum # Could just Kruskal-Walis to see if any of the # experiments has different expression. # Remember that first column is the probe names kruskal.test(ge[,2:7]) Kruskal-Wallis rank sum test data: ge[,2:7] Kruskal-Wallis chi-squared = 58.4336, df = 5, p-value = 2.56e-11 # Highly significant, but it doesn’t tell us which # Have a look apply(ge[,2:7],2,median) Exp1 Exp2 Exp3 Exp4 Exp5 Exp6 38.6 40.4 40.1 36.7 36.5 37.7 # MAD is the robust equivalent of standard deviation # Median Absolute Deviation, that is the median distance # from the median. apply(ge[,2:7],2,mad) Exp1 Exp2 Exp3 Exp4 Exp5 Exp6 52.33578 53.96664 53.81838 50.11188 50.11188 51.00144 # The largest difference appears to be between Exp2 and # Exp5 but our measure of variance is large (although # we haven’t scaled it for number of samples yet). apply(ge[,2:7],2,mad)/sqrt(22809) Exp1 Exp2 Exp3 Exp4 Exp5 Exp6 0.346534 0.357332 0.356351 0.331809 0.331809 0.337699 # Max difference is 3.9, so plausible that we could # detect an effect. # Have many genes and experiments, so most appropriate # to use Friedman test. # For arcane reasons, we have to tell R to interpret data # as a matrix. friedman.test(as.matrix(ge[,2:7])) Friedman rank sum test data: as.matrix(ge[,2:7]) Friedman chi-squared = 2462.051, df = 5, p-value < 2.2e-16 # Much more significant result than Kruskal-Walis test friedman.test(as.matrix(ge[2:7]))$p.value [1] 0 # Compare power by looking at subset of data kruskal.test(ge[1:1000,2:7]) Kruskal-Wallis rank sum test data: ge[1:1000, 2:7] Kruskal-Wallis chi-squared = 7.4089, df = 5, p-value = 0.1920 friedman.test(as.matrix(ge[1:1000,2:7])) Friedman rank sum test data: as.matrix(ge[1:1000, 2:7]) Friedman chi-squared = 155.5744, df = 5, p-value < 2.2e-16