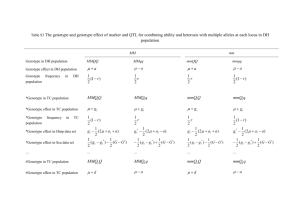
PBG/MCB 622 11/26/2012 Exercise Association Mapping using
advertisement

PBG/MCB 622 11/26/2012 Exercise Association Mapping using TASSEL Open TASSEL, Load Genotypic and Phenotypic data going to Data -> Load -> “I will make my best guess and try” Calculation of Linkage disequlibrium: We will calculate LD for chromosome 1 only. First we will filter our dataset selecting only markers on chromosome 1, with a minor allele frequency higher than 5% and less than 5% of missing data. Select the genetotypic data (“Genotype_AM”) and go to Data->Sites-> Minimum count: 97 Minimum frequency: 0.05 Start Position:0 End Position: 236 Check “Remove minor SNP states” Filter Select the filtered database and go to Analysis->Linkage disequilibrium You can plot the results selecting the output LD file and going to Results->LD plot Calculation of Population Structure Using Principal Components Analysis (Q Matrix) Make sure TASSEL is in Data mode. Highlight the genotype and click Site. Set the minimum frequency to 0.05, Minimum count to 97 and have “Remove minor SNP status”. Click Filter. Numericalization: Highlight the filtered genotype and click Transform. Use the default option of “Collapse non major alleles.” Click Create data set. Imputation of missing values: Highlight the numerical genotype and click Transform and then click Impute Tab. Use the default options. Click Create data set. PCA: Highlight the imputed numerical genotype, click Transform, and then click PCA Tab. Change the default option to “Components=3” by choosing Components and type 3 in the text box. Click Create data set. You can plot the results selecting the output PCA file and going to Results->Chart Calculation of the Kinship Matrix (K Matrix) using SNP data Remove monomorphic sites: Highlight the genotype and click Site in Data mode. Set the threshold on MAF to 0.05, Minimum count to 97, check “Remove minor SNP status,” then click Filter. Estimate kinship: Highlight the filtered genotype and click Kinship in Data mode. A kinship matrix will be added to the data tree under Matrix category. Association analysis using GLM (Least squares fixed effects linear model) 1) Naïve Model: Flowering time = Marker effect + residual Remove monomorphic sites: Highlight the genotype and click Site in Data mode. Set the threshold on MAF as 0.05, Minimum count to 97, then click Filter. Joining data: Highlight the two data sets (Filtered genotype and phenotype) by holding the Control key while selecting the individual data. Then click Intersection (∩) Join on Data mode to create a combined data set. Association analysis: Highlight the joint data set then click GLM in Analysis mode to perform association analysis. Two reports will be added to the data tree. Visualize results by selecting the “GLM_Marker_test” results file and clicking Results-> Manhattan Plot 2) Q Model: Flowering time = Population Structure+Marker effect + residual Remove monomorphic sites: Highlight the genotype and click Site in Data mode. Set the threshold on MAF as 0.05, Minimum count to 97, then click Filter. Joining data: Highlight the three filtered data sets (“PC_genotype_for_AM”, Filtered genotype and phenotype) by holding the Control key while selecting the individual data. Then click Intersection (∩) Join on Data mode to create a combined data set. Association analysis: Highlight the joint data set then click GLM in Analysis mode to perform association analysis. Two reports will be added to the data tree. Visualize results by selecting the “GLM_Marker_test” results file and clicking Results-> Manhattan Plot Association analysis using MLM (Mixed linear model: Individuals and the residual are fit as random effects. The other terms are treated as fixed effects.) 3) Q+K Model: Flowering time = Population Structure+ Marker effect + Individuals + residual Association analysis: Highlight the joint data set created for the Q model (Phenotype+genotype+PCA) and the Kinship Matrix. Click MLM in Analysis mode. Apply all the default settings. Visualize results by selecting the “GLM_Marker_test” results file and clicking Results-> Manhattan Plot