Final Project Instruction
advertisement

CAP4770 – Introduction to Data Mining
Final Project Instruction
Data:
We use a gene data set as our data for the final project. This data set is in
attributes-in-rows format, comma-separated values. It can be downloaded by
following this link:
http://users.cis.fiu.edu/~lli003/teaching/hw-sol/finalproject_datafiles.zip
Username/Password: CAP4770/student
The zip file contains three files:
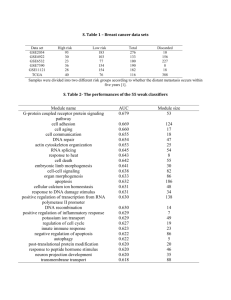
train.csv: training data, consisting of 69 instances with 7,070 attributes.
train_class.txt: training classes, corresponding to the true labels for each
instance in training data in the order. There are 5 classes in total, MED,
MGL, RHB, JPA and EPD.
test.csv: test data, consisting of 112 unlabeled instances with 7,070
attributes.
Goal:
Learn the best classifier from the training data and use it to predict the classes
for test data.
Due Date:
December 6th, 2012
Submission:
1. Project report, describing how to establish your classifier step by step.
Specifically, your report should include:
1) How to do data cleaning;
2) How to do feature selection;
3) How to train classifier.
2. Predicted result (in YourPatherId.txt file, one class per line in uppercase, as
the same order of the test data)
3. Make sure that all the files you submit are zipped into one single file,
named as “CAP4770_finalproject_firstname_lastname_patherid”.
Important hints:
1. The training and testing data are all in the format of attribute-in-row.
Probably you need to transform the data into the format of attribute-incolumn so that the data can be appropriately fed into Weka.
2. You need to do some preprocessing, e.g., feature selection to select N
attributes that are more important and predictive, by using Weka or writing
a java program.
3. You can use any classification method available in Weka or some other
tools, as long as you explain well why you choose it in your report.
4. You can use ensemble classification to get more reasonable classifier;
however, you need to consider carefully from technical perspective how to
integrate different classifiers together to get better results.
5. It is better if I can reconstruct your classifier with my own environment;
therefore, you’d better provide me a specific instruction in your report on
how to build your own classifier.
6. More accurate your classifier is, more detailed explanation your report has,
more points you get.
The following steps suggest one way of finding the best classifier. Note that
this is just one way for doing the project. You can definitely make improvements
or use other ways to get a classifier with higher accuracy.
1. Data Cleaning
Threshold both train and test data to a minimum value of 20, maximum of 16,000.
2. Selecting top genes (i.e., attributes) by class
You can use any feature selection methods from Weka.
o Here is one method:
remove from train data genes with fold differences across
samples less than 2; (Note that Fold difference is defined as
(max-min)/2 where. max and min are the maximum and
minimum values of the gene expression for all the instances,
respectively. A fold difference of less than 2 means that across
the samples, the gene value does not change much and as such,
cannot influence the class significantly.)
for each class, generate subsets with top
2,4,6,8,10,12,15,20,25, and 30 top genes with the highest Tvalue
Optional: for each class, select top genes using
highest absolute T-value (i.e. also include genes with high
negative T-value) (Note that T-value is calculated as follows:
( Avg1 Avg2 )
( 12 / N1 22 / N 2 )
where Avg1 is the average for one class
across the gene sample and Avg2 is the average for the other 4
classes. Stdev1 is the standard deviation for one class and
Stdev2 is the standard deviation for the other classes. Similarly,
N1 is the number of samples that have the class whose T-value
we are interested in, and N2 is the number of samples that
does not have the T-value that we are interested in. )
for each N=2,4,6,8,10,12,15,20,25,30 combine top genes for each class into
one file (removing duplicates, if any) and call the resulting file
train_topn.csv
Add the class as the last column, remove sample no, transpose each file to
“attributes-in-columns” format and convert it to .arff format.
3. Find the best classifier/best gene set combination
You can use any classifiers you want. For example, you can use:
NaiveBayes
J48
IB1
IBk (for each value of K=2, 3, 4)
one more Weka classifier of your choice -- that can work with multiclass
data.
a. For each classifier, using default settings, measure classifier accuracy on the
training set using previously generated files with top
N=2,4,6,8,10,12,15,20,25,30 genes.
For IBk, test accuracy with K=2, 3 and 4.
b. Select the model and the gene set with the lowest cross-validation error.
Optional: once you found the gene set with the lowest cross-validation error,
you can vary 1-2 additional relevant parameters for each classifier to see if the
accuracy will improve. E.g. for J4.8, you can vary reducedErrorPruning and
binarySplits
c. Use the gene names from best train gene set and extract the data
corresponding to these genes from the test set.
d. Convert test set to genes-in-columns format.
e. Add a Class column with “?” values as the last column
4. Generate predictions for the test set
You should now have the best train file, call it train.bestN.csv, (with 69
samples and bestN number of genes for whatever bestN you found) and a
corresponding test file, call it test.bestN.csv, with the same genes and 112 test
samples. The train file will have all Class values while the test file Class column
will have only “?”.
a. Convert test file to arff format (you should already have .arff for train file
from Step 3).
Important: In Weka, the variable declarations should be exactly the same for
test and train file. To achieve that, change the Class entry in test.bestN.arff
header section to be the same as in train file, i.e.
@attribute Class {MED,MGL,RHB,EPD,JPA}
b. Use the best train file and the matching test file and generate predictions
for the test file class.
5. Write a paper describing your effort.
The above steps are just one way to classify the test data. You can come up
with your own solution, as long as the procedure is reasonable.