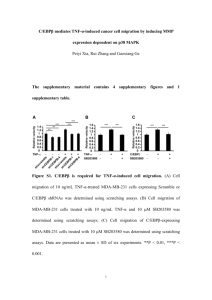
Synthesis and characterization of MC3482 Chemistry The synthetic
advertisement

Synthesis and characterization of MC3482 Chemistry The synthetic pathway to compound MC3482 is depicted in Schema 1. The commercially available (L)-Z-Lys(Boc)-OH has been coupled with aniline in the presence of the N-ethyl-N'-(3dimethylaminopropyl)carbodiimide hydrochloride (EDCI)/hydrozybenzotriazole (HOBt) coupling system and triethylamine(TEA) in dry dichloromethane(DCM) to provide the intermediate 1. The Boc(tert-butoxycarbonyl) deprotection of 1 has been performed with 4NHCl (1,4-dioxane solution) in dry dichloromethane to furnish the hydrochloride 2, which has been treated with the commercially available (L)-Z-Glu-OtBuin the presence of the EDCI/HOBt coupling system and triethylamine in dry DMF to give the intermediate 3. The final compound MC3482 has been obtained by Boc deprotection of 3 through the use of trifluoroacetic acid (TFA) in dry dichloromethane at room temperature (Schema 1). Schema 1.a 1 a Reagents and conditions: (a) aniline, EDCI, HOBt, TEA, dry DCM, rt, 16h, 84%; (b) HCl 4N in dioxane, dry DCM, rt, 24 h, 92%; (c) (L)-Z-Glu-OtBu, EDCI, HOBt, TEA, dry DMF, rt, 16h, 79%; TFA, dry DCM, rt, 24 h, 96%. MC3482, (S)-2-(((benzyloxy)carbonyl)amino)-5-(((S)-5-(((benzyloxy)carbonyl)amino)-6- oxo-6-(phenylamino)hexyl)amino)-5-oxopentanoic acid: Recrystallization system: CH3CN; mp 148-150 °C.1H-NMR (DMSO) 1.24-1.38 (m, 4H, CH2CH2CH2NHCO), 1.63 (m, 2H, CH2CH2CH2CH2NHCO), 1.76 (m, 1H, CHHCH2CONH), 1.95 (m, 1H, CHHCH2CONH), 2.13 (m, 2H, CH2CH2CONH), 3.00 (m, 2H, CH2CH2CH2CH2NHCO), 3.93 (m, 1H, NHCHCOOH), 4.11 (m, 1H, NHCHCONHPh), 5.03 (s, 4H, 2 x OCOCH2Ph), 7.04 (m, 1H, benzene ring), 7.28-7.36 (m, 12H, benzene rings), 7.54-7.60 (m, 4H, benzene rings and 2 x NHCOOBn), 7.80 (m, 1H, CH2NHCOCH2), 9.99 (s, 1H, CONHPh), 12.20 (s, 1H, COOH). Figure S1. Inhibition of SIRT5 desuccinylasic activity by MC3482. MDA-MB-231 WT cells were cultured for 24 h in the presence or absence of MC3482 at the concentrations reported in the figure. SIRT5 desuccinylating activity was measured as reported under Materials and Methods. Data are representative of at least 3 separate experiments. *Significantly different from WT untreated cells. Significance was set at P<0.05. Figure S2. SIRT5 inhibitor MC3482 does not inhibit SIRT1 and SIRT3 activity. MDA-MB-231 WT cells were cultured for 24 h in the presence or absence of 50 μM MC3482. SIRT1 and SIRT3 activity was measured as reported under Materials and Methods. Data are representative 2 of at least 3 separate experiments. Figure S3. GLUD1 immunoprecipitation and activity in WT and SIRT5 clones. (A) MDA-MB231 WT cells in the presence or absence of MC3482, as well as SIRT5+ and SIRT5- clones were immunoprecipitated with an anti-GLUD1 antibody and blotted for SIRT5 (left side) or GLUD1 and SIRT3 (right side). Densitometric analysis of the bands was determined as under Materials and Methods and is reported below each blot. (B) GLUD1 activity in MDA-MB-231 WT cells in the presence or absence of MC3482, as well as SIRT5+ and SIRT5- clones was determined in not treated conditions (nt) and in the presence of the GLUD1 inhibitors hexachlorophene or dimethyl-α-ketoglutarate as reported under Materials and Methods. Data are representative of at least 3 separate experiments. *Significantly different from nt cells. Significance was set at P<0.05. Figure S4. Ammonia production after glutamine withdrawal.MDA-MB-231 and C2C12 WT cells in the presence or absence of MC3482, as well as SIRT5+ and SIRT5- clones were cultured in the absence of glutamine for the times indicated. Ammonia levels were measured in the culture medium every other day as reported under Materials and Methods. Data are representative of at least 3 separate experiments. Figure S5. Interaction between SIRT5 and catalytic domain of GLS (cGLS).Molecular surface representation in the frontal view (A) and lateral view (B) showing that SIRT5 (purple) precisely fits into the cavity of cGLS tetramer (green). These images were obtained using Chimera 1.8 molecular graphics program. 3 Figure S6. Catalytic domain of GLS (cGLS) and its possible interaction with SIRT5.(A) Molecular surface representation of the cGLS structure. The active site cleft is shown in blue. Lys320 (yellow stick and transparent surface) and Lys245 amino acids (magenta stick and transparent surface) are highly exposed and resides on opposite sides on surface of the GLS active site. (B) SIRT5 catalytic pocket (transparent purple ribbon) facing the active site of GLS (molecular surface representation). The residues located at the entrance of the SIRT5 catalytic pocket are Phe223, Leu227, Val254 (red stick), whereas Arg105 and Tyr102 (orange stick) are located on the bottom of the catalytic pocket where the interaction that stabilizes the succinyl group takes place. Lys320 and Lys245 are also shown and their location suggests that they may be accessible to the SIRT5 catalytic pocket. The 3D model image was obtained using Chimera1.8 molecular graphics program. Figure S7. Ortholog sequences alignment of cGLS. Sequences of human, mouse, rat, and bacteria GLS. The code UniProt/Swiss-Prot and the numbering of the residues in the sequence are also shown on the left side. Percentage of identity among the sequences is indicated on the right side. The red box indicates Lys245 that is conserved in the sequence of mouse and rat, in other isoforms of cGLS, and in some bacteria. Lys245 is localized between Gly at the -1 position (conserved in all species) and Val at the +1 position. The blue box indicates Lys320 preservation, equivalent of Lys253 in GLS2 [glutaminase 2 (liver, mitochondrial)] of mouse identified as a succinylation-lysine site by Park et al.28 *=100% match among residues; :=high similarity (> 75%); .=average similarity(50% -75%). The alignment was obtained using the Clustal Omega program. 4 Figure S8. LC3 fluorescent puncta increase after SIRT5 silencing or inhibition and with NH4Cl. MDA-MB-231 WT cells were cultured for 1 or 4 d in the presence or absence of MC3482 and 2 mM NH4Cl. LC3 puncta accumulation was measured as reported under Materials and Methods. Data are representative of at least 3 separate experiments. *Significantly different from WT untreated cells. Significance was set at P<0.05. 5