ChIP protocol
advertisement

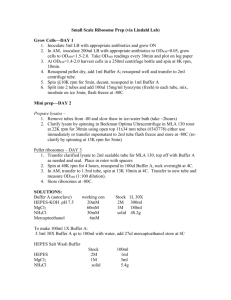
ChIP from brain tissue Protocol updated Dec. 2010 AW Pool the frozen striata from 3-5 mice **For 4-5 ChIPs in duplicate, ideally this is both sides. For cells start with 10-20 million cells/IP. Add 500µL of 1% formaldehyde in PBS to brains in a 2mL glass homogenizer at RT. Dounce with 90 strokes, move to eppendorf tube and leave the samples at RT to fix for 10’ while rotating. For cells, just add Formaldehyde to cells at 1% and rotate for 10’ Quench fixation by adding 80µL of 1.25M glycine to 500µL volume (200mM final concentration) for 5 min at RT on rotator. Pellet cells by spinning 5’ at 7500RPM, wash 3X 1mL ice cold PBS with protease inhibitors, then spin down again and resuspend in 500µL lysis buffer (50mM Tris-HCl pH 8.0, 150mM NaCL, 5mM EDTA, 1% v/v NP-40, 0.5% w/v sodium deoxycholate, complete protease inhibitors). Lyse for 15’ rotating at 4 degrees. **If there is less tissue, the sonication will need to be stronger/longer. For cells you will need a tissue scraper to scrape them off the plate before spinning down. In terms of volumes, 500µL is a good volume for sonication – hard to go lower without foaming. But you need enough cells in that volume. Most likely this is about 200 million cells in 500µL. Hold the tube in a small bucket of ice in your hand and sonicate with Misonix sonicator 5 times 15 seconds at power 6.0 equivalent to ~30 watts. Rest on ice 15 seconds in between each round. If the output does not reach 27-30 watts, raise the power to 6.5 or 7. Goal is to get genomic fragments of 400-2000bp. **Can sonicate again after the test if needed. Remove insoluble material by spinning 14,000RPM (20,000 RCF) for 10’ at 4 degrees. Spec the chromatin. If you dilute 2uL in 100uL water, except OD260 about 0.2, which is 500ng/µL. By my calculation from what I had read in other papers I thought this was correct. However when I actually did the ChIP, it was very hard to get a good enough yield with these kind of spec values. Instead, I used at least twice this much (so 50µg by this calculation) or in the optimal ones I did, 4-5 times this much starting material. For each IP use 25µg (50-125µg) chromatin. For the OD value above this is 50µL, meaning that there is enough chromatin for 10 total pulldowns in each 500µL sonicated sample. Ideally this is four antibodies in duplicate with duplicate input samples (=10). Don’t forget to save 2 aliquots of cleared chromatin for the input samples. The way these are processed is described below. Take that volume of chromatin and dilute in 500µL lysis buffer with protease inhibitors. Swell 0.25g protein A sepharose (GE cat # 17-0780-01) in 2 x 50mL water. Allow to settle or spin 3’ at 500 x g. Block beads in 1mL 3% BSA in lysis buffer for 1 hour at RT on the rotator. Wash 2X lysis buffer (equal volume to bead bed) spinning no more than 500 x g each time (2500RPM in eppy fuge). Store beads at 4 degrees. Preclear with 25uL of 50% protein A sepharose /PBS slurry for 1-2 hrs. at 4 degrees. Set aside two tubes of 10% of total chromatin and preclear sample as Input DNA. This reenters protocol at the crosslink reversing step below. After preclear, spin out beads at 7500RPM for 5’. Move supe to new tube. For each IP add 5ug of the antibody (about 5uL usually) and go overnight at 4 degrees rotating. Collect complexes by adding 50uL of 50% protein A sepharose/PBS slurry for 1 hr at 4 degrees. Spin down 3’, 500 x g (2500 RPM in Eppy fuge). Wash 2 x 5 min room temp lysis buffer, 2 x ChIP wash buffer (50mM Tris-HCl 8.5, 500mM LiCl, 5mM EDTA, 1%NP-40, 1% sodium deoxycholate), 2 x TE. 500µL each wash. Spin down 3’, 500 x g (2500 RPM in Eppy fuge). **Careful on the last wash not to remove the beads. Elute in 100uL elution buffer (50mM Tris-HCl 8.0, 1% SDS, 1mM EDTA) at 65 degrees for 10 min followed by vortexing for 15 sec. Spin down 7500 RPM for 5’ and transfer supe to new tube. The input samples reenter the protocol here. To each add 100uL elution buffer and then go on in parallel. Add 4µL 5M NaCl (200mM final) and incubate at 65 degrees overnight to reverse crosslinks. Add 1uL 10mg/mL RNAseA and incubate 1 hour at RT. Add 1uL 20U/mL (Qiagen) proteinase K and incubate for 30’ at 55 degrees. Prepare a phase lock light tube (5 prime 2302800) by spinning top speed in Eppy fuge 30sec. Add 100uL sample and 100uL buffer saturated phenol/chloroform/IAA. Mix without vortexing. Spin top speed 5’. Remove top phase above the gel to a new tube. Add 2uL glycogen (Invitrogen) and 250µL 100% EtOH. Precipitate at -80 for 2 hours to overnight. Spin down 30’ at 4 degrees. Wash 1X 70% EtOH and spin again 5’ 4 degrees. Resuspend in 30µL Tris pH 8.0. Aliquot and store at -80. Limit freeze/thaw. Limiting the freeze-thaw really matters. In our protocol you need 3.25µL/PCR for each primer pair, so this will give you about 8 targets, put in 8 tubes. What I recommend is that you start this protocol with some specific number of cells, test your chromatin recovery by spec, then take an input sample through the final stages and test recovery by PCR. When you PCR from your input sample for your gene of interest you expect the Cts to be in the 20-25 range. You can run a dilution series of the input to test your primer efficiency at the same time. Finally run the input material on a gel to ensure that it is a smear of about 200-1000bp, you are looking for an average sonicated fragment size of 500bp. I can show you some pictures so you can see what it should look like and what it should not look like. In the meantime, overexpress your protein of interest, fix the cells this way, and test the IP with your antibodies to see how efficient they are for the pull-down. You can test that by western comparing immunoreactivity in cells before IP, supe after IP, and pellet after IP. It is ideal if you can pull down with one Ab and blot back with another – particularly of another species to avoid the light and heavy chains (what MW is cut? LC is 25kd, HC is 55 so hard to see at those MWs if you can’t switch species). Hope this helps!! Good luck.