Supplementary Figures (docx 483K)
advertisement

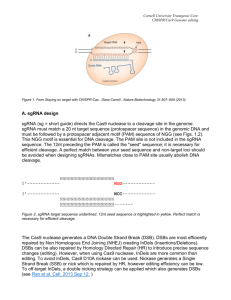
Supplementary Information: CRISPR-mediated activation of latent HIV-1 expression Prajit Limsirichai1, *, Thomas Gaj2, * and David V. Schaffer2, 3, 4, 5 1 Department of Plant and Microbial Biology, University of California, Berkeley, Berkeley, California 94720, USA 2 Department of Chemical and Biomolecular Engineering, University of California, Berkeley, Berkeley, California 94720, USA 3 Department of Bioengineering, University of California, Berkeley, Berkeley, California 94720, USA 4 Department of Cell and Molecular Biology, University of California, Berkeley, Berkeley, California 94720, USA 5 Helen Wills Neuroscience Institute, University of California, Berkeley, Berkeley, California 94720, USA * These authors contributed equally to this work Correspondence and requests for materials should be addressed to D.V.S. (schaffer@berkeley.edu) ACTGGAAGGGCTAATTCACTCCCAAAGAAGACAAGATATCCTTGATCTGTGGATCTA CCACACACAAGGCTACTTCCCTGATTGGCAGAACTACACACCAGGGCCAGGGGACA GATATCCACTGACCTTTGGATGGTGCTACAAGCTAGTACCAGTTGAGCCAGATAAGA TAGAAGAGGCCAATAAAGGAGAGAACACCAGCTTGTTACACCCTGTGAGCCTGCAT GGGATGGATGACCCGGAGAGAGAAGTGTTAGAGTGGAGGTTTGACAGCCGCCTAGC ATTTCATCACGTGGCCCGAGAGCTGCATCCGGAGTACTTCAAGAACTGCTGATATCG AGCTTGCTACAAGGGACTTTCCGCTGGGGACTTTCCAGGGAGGCGTGGCCTGGGCG GGACTGGGGAGTGGCGAGCCCTCAGATCCTGCATATAAGCAGCTGCTTTTTGCCTGT ACTGGGTCTCTCTGGTTAGACCAGATCTGAGCCTGGGAGCTCTCTGGCTAACTAGGG AACCCACTGCTTAAGCCTCAATAAAGCTTGCCTTGAGTGCTTCAAGTAGTGTGTGCC CGTCTGTTGTGTGACTCTGGTAACTAGAGATCCCTCAGACCCTTTTAGTCAGTGTGG AAAATCTCTAGCAG Supplementary Fig. 1. Long-terminal repeat (LTR) sequence from the HIV-1 subtype B strain HXB2 (NCBI accession number: K03455.1). sgRNA target sites are highlighted yellow. Positions of variation between the reference LTR and the sequence used for the design and validation of the sgRNA are indicated in red. 2 Supplementary Fig. 2. “Tiling” multiple sgRNAs across the HIV-1 LTR promoter does not dramatically enhance the efficiency of dCas9-VP64-mediated gene activation. Fold-increase in the percentage of EGFP positive HEK293T cells after transfection with 20 ng of EGFP reporter plasmid, 90 ng of dCas9-VP64 expression vector, and 90 ng of sgRNA expression vector. For tiling experiments, the amount of sgRNA-encoded DNA was kept constant so that each transfection contained 90 ng of total sgRNA. Data normalized to EGFP expression in cells transfected with sgRNA 3. Only sgRNAs 4 and 7 induced a significant increase in gene activation compared to sgRNA 3. See Fig. 1d for the activity of each individual sgRNA. EGFP expression was measured 48 hr after transfection. Error bars indicate s.d. (n = 3; *p-value < 0.05; Student’s t-test). 3 Supplementary Fig. 3. Tiling multiple sgRNAs across the HIV-1 LTR promoter does not significantly increase the efficiency of SAM-mediated gene activation. Fold-increase in the percentage of EGFP positive HEK293T cells after transfection with 20 ng of EGFP reporter plasmid, 60 ng of dCas9-VP64 expression vector, 60 ng of MS2-p65-HSF1 expression vector, and 60 ng of sgRNA expression vector. For tiling experiments, the amount of sgRNA-encoded DNA was kept constant so that each transfection contained 60 ng of total sgRNA vector. Data normalized to EGFP expression in cells transfected with sgRNA 3. See Fig. 1d for the activity of each individual sgRNA. EGFP expression was measured 48 hr after transfection. Error bars indicate s.d. (n = 3; p-values from Student’s t-test). 4 Supplementary Fig. 4. Co-transfection of SAM with a library of non-specific sgRNA does not lead to a significant increase in gene activation from the HIV-1 LTR. Fold-increase in the percentage of EGFP positive HEK293T cells after transfection with 20 ng of EGFP reporter plasmid, 60 ng of dCas9-VP64 expression vector, 60 ng of MS2-p65-HSF1 expression vector, and 60 ng of an empty sgRNA expression cassette, or a library1 of sgRNAs designed to target the proximal promoter of each human RefSeq coding isoform. EGFP expression was normalized to cells transfected with the empty sgRNA expression cassette and measured 48 hr after transfection. Error bars indicate s.d. (n = 3; p-value from Student’s t-test). 5 Supplementary Fig. 5. Tumor necrosis factor alpha (TNF-α) stimulation of J-Lat 9.2 and JLat 10.6 cells. Distribution histograms generated by flow cytometry indicating the percentage of GFP positive naïve J-Lat 9.2 (upper) and J-Lat 10.6 (lower) cells following stimulation with increasing amounts of TNF-α. GFP expression was measured 24 hr after treatment. 6 Supplementary Fig. 6. Prostratin treatment of J-Lat cells expressing SAM is not as effective at stimulating viral gene expression as combination treatment with SAHA. Percentage of GFP positive J-Lat 9.2 (upper) and J-Lat 10.6 (lower) cells stably expressing SAM with sgRNAs 1 to 7 and treated with either 4 µM SAHA only, 2 µM prostratin only, or 4 µM SAHA plus 2 µM prostratin. “Empty” indicates J-Lat cells expressing SAM with an empty sgRNA expression cassette. Dotted line indicates the percentage of GFP positive naïve J-Lat cells after treatment with 20 ng/µL of TNF-α. Cells were treated 17 days after infection, and GFP expression was measured 24 hr after treatment. Error bars indicate standard deviation (n = 3). 7 5’ LTR sgRNA 1 CACCGACAAGATATCCTTGATCTG 3’ LTR sgRNA 1 AAACCAGATCAAGGATATCTTGTC 5’ LTR sgRNA 2 CACCGATTGACAGAACTACACACC 3’ LTR sgRNA 2 AAACGGTGTGTAGTTCTGTCAATC 5’ LTR sgRNA 3 CACCGTCAGATATCCACTGACCTT 3’ LTR sgRNA 3 AAACAAGGTCAGTGGATATCTGAC 5’ LTR sgRNA 4 CACCGCTACAAGGGACTTTCCGCT 3’ LTR sgRNA 4 AAACAGCGGAAAGTCCCTTGTAGC 5’ LTR sgRNA 5 CACCGGCGTGGCCTGGGCGGGACT 3’ LTR sgRNA 5 AAACAGTCCCGCCCAGGCCACGCC 5’ LTR sgRNA 6 CACCGGTTAGACCAGATCTGAGCC 3’ LTR sgRNA 6 AAACGGCTCAGATCTGGTCTAACC 5’ LTR sgRNA 7 CACCGCCCGTCTGTTGTGTGACTC 3’ LTR sgRNA 7 AAACGAGTCACACAACAGACGGGC Supplementary Fig. 7. Primer sequences used to construct the sgRNA used in this study. Bases complementary to the LTR are yellow highlighted. 8 REFERENCES 1. Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPRCas9 complex. Nature 517, 583-588 (2015). 9