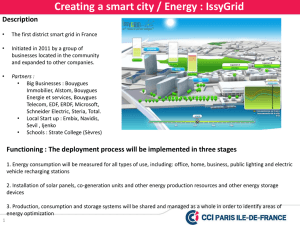
Supplementary Data - European Heart Journal
advertisement

-1- Online supplementary data Towards a Clinical Use of Human Embryonic Stem Cell-Derived Cardiac Progenitors: A Translational Experience Philippe Menasché1,2,3*, Valérie Vanneaux,4,5,6 Jean-Roch Fabreguettes7, Alain Bel1,3, Lucie Tosca8, Sylvie Garcia9, Valérie Bellamy3, Yohan Farouz2,3, Julia Pouly1, Odile Damour10, Marie-Cécile Périer11 Michel Desnos2,3,12, Albert Hagège2, 3, 12 , Onnik Agbulut13, Patrick Bruneval2,11,14, Gérard Tachdjian8, Jean-Hugues Trouvin15,16, Jérôme Larghero4,5,6. 1 Assistance Publique-Hôpitaux de Paris, Hôpital Européen Georges Pompidou, Department of Cardiovascular Surgery, Paris, France. 2 University Paris Descartes, Sorbonne Paris Cité, F-75475, Paris, France. 3 INSERM U 633, Hôpital Européen Georges Pompidou, Paris, France. 4 Assistance Publique-Hôpitaux de Paris, Hôpital Saint-Louis, Cell Therapy Unit and Clinical Investigation Center in Biotherapies (CBT501), Paris, France. 5 University Paris Diderot, Sorbonne Paris Cité, F-75475 Paris, France. 6 INSERM UMRS940, Institut Universitaire d’Hématologie, Hôpital Saint-Louis, Paris, France. 7 Assistance Publique-Hôpitaux de Paris, Central Pharmacy, Clinical Trials Department, Paris, France. 8 Assistance Publique-Hôpitaux de Paris, University Paris Sud, Histology-Embryology- Cytogenetics, Hôpitaux Universitaires Paris Sud, 92141 Clamart, France. 9 Unité de Biologie des Populations Lymphocytaires, Department of Immunology, Institut Pasteur, CNRS-URA 1961, Paris, France. 10 Tissues and Cells Bank, Edouard Herriot Hospital, Lyon, France. -211 INSERM U 970, Hôpital Européen Georges Pompidou, Paris, France. 12 Assistance Publique-Hôpitaux de Paris, Hôpital Européen Georges Pompidou, Department of Cardiology, Paris, France. 13 UPMC University Paris 6, Sorbonne Universités, Department of Aging, Stress and Inflammation, Paris, France. 14 Assistance Publique-Hôpitaux de Paris, Hôpital Européen Georges Pompidou, Department of Pathology, Paris, France. 15 University Paris Descartes, School of Pharmacy, Paris, France. 16 Assistance Publique-Hôpitaux de Paris, Central Pharmacy, Pharmaceutical Innovation Department, Paris, France. *To whom correspondence should be addressed : Department of Cardiovascular Surgery, Hôpital Européen Georges Pompidou, 20, rue Leblanc, 75015 Paris, Phone : 33 1 56 09 36 22; Fax : 33 1 56 09 32 61; philippe.menasche@egp.aphp.fr. France. -3- SUPPLEMENTAL MATERIALS AND METHODS Procurement, derivation and initial testing of the I6 ESC line. The I6 cell line was obtained from a leftover zygote after an in vitro fertilization procedure at the Rambam Medical Center (Israël). This zygote was donated by a couple after it had signed consent forms. The zygote was cultured to the blastocyst stage using Cook media (Cook, Queensland, Australia). After zona pellucida digestion by Tyrode’s acidic solution (Sigma, St Louis, MO, USA), the blastocyst was incubated for 30 minutes with anti-human whole antiserum antibody. The embryo was washed three times using Gibco® Dulbecco's Modified Eagle Medium (DMEM, Life Technologies, Grand Island, NY, USA) and then incubated with 0.5% Guinea pig complement (Gibco) for about 20 minutes. At the end of the incubation, the embryo was washed three times using DMEM and then placed onto mitotically inactivated mouse embryonic fibroblasts (MEF) derived on-site from Imprinting Control Region (ICR) mice (Harlan, Israel). Cells were grown in a culture medium consisting of 80% Knock-Out (KO)-DMEM, supplemented with 20% defined fetal bovine serum (FBS, HyClone, Logan, UT, USA), 1mM L-glutamine, 0.1 mM β–mercaptoethanol, 1% non-essential amino acid stock (all from Invitrogen™, Life Technologies, Carlsbad, CA). During the first few passages, the cells were passaged mechanically using 27 gauge syringes. Later, the cells were passaged every four to six days using 1mg/ml type IV collagenase (Invitrogen™). A MCB was then prepared after cells were frozen in liquid nitrogen using a freezing solution consisting of 10% dimethyl sulfoxide (DMSO, Sigma, St Louis, MO, USA), 20% FBS and 80% KO-DMEM. Genetic stability : The human ESC line I6 was tested for karyotypic stability 7 months after its derivation (at passage 29). This analysis was conducted at the Cytogenetic Unit of Rambam Health Care Campus. G-band standard staining (Giemsa, Merck, Darmstadt, Germany) was used for chromosome visualization. The karyotypes were analyzed and reported according to -4- the "International System for Human Cytogenetic Nomenclature" (ISCN). One-hundred metaphases were examined and the cell line was found as normal 46, XY. Sterility: Different batches were tested for mycoplasma, using a commercial kit (Biological Industries, Kibbutz Beit-Haemek, Israel) or Polymerase Chain Reaction (PCR) according to the protocol provided by the British National Cell Bank and were found negative. Viral testing : No evidence for murine leukemia viruses (MuLVs) transmission from MEFs to the I-6 line was found1. The HSV-1, HSV-2, CMV and HBV viruses, tested by nested PCR at the Rambam Medical Center Virology Laboratory were also found negative. Such was also the case for the HIV virus tested by PCR at the Sheba Medical Center Virology Laboratory. hESCs features: The pluripotency of the cell line was confirmed by its positive immunostaining for undifferentiated stem cell-associated genes (SSEA-3, SSEA-4, Oct 4, tumor rejection antibody [TRA]-1-60, TRA-1-81), the formation of embryoid bodies and the generation of teratomas following cell injection into the rear leg muscle of 4-week-old male SCID-beige mice.2 Cell expansion. Cell expansion was performed in a similar way in the two laboratories (Hôpital Saint-Louis and MAbgène). Briefly, thawed cells (viability > 50%) were washed in10 mL of Nutristem™ hESC XF (Biological Industries, Kibbutz Beit-Haemek, Israel) and centrifuged at 800 rpm for 4 minutes. The pellet was then collected and suspended in Nutristem™. Cells were seeded onto 15cm dishes (B15) previously seeded with clinical-grade irradiated human foreskin fibroblasts (Tissue Bank, Hôpital Edouard Herriot, Lyon) at 37°C with 5% CO2 and the medium was changed daily. When I6 ES cells reached 80-90% confluence, cells were dissociated from the feeder fibroblasts by treatment with collagenase (1mg/mL, Collagenase NB6, GMP grade, batch #22540105, Nordmark, Uetersen, Germany) during 40±10 minutes. The I6 ESC colonies -5- were removed from the surface of dishes by collecting the supernatant. After the washing step of dishes, the pooling of the supernatant and the washing medium and centrifugation of the pool contents at 800 rpm for 4 minutes, the I6 cells were replated onto fibroblast-pre-seeded dishes at the usual 1:5 split ratio. Preliminary experiments have estimated that, at this stage, the number of cells present in a B15 dish is approximately 10 million. All calculations have thus been based on this metric. Care was taken to passage the cells in small clumps to enhance their viability. At the time of banking, cells were cryopreserved in FBS (HyClone, Thermo-Fisher, Illkirch, France) and 10% DMSO (B.Braun Medical, Boulogne, France). Back-and forth transfers of the cells between the two sites (Hôpital Saint-Louis and MAbgène) were performed under well controlled transport conditions in vapour nitrogen dry shippers. As mentioned in the main manuscript, for the pilot phase I clinical trial, it has been decided to pool the master and the working cell banks together. A two-tiered banking system will be established later on depending on the outcome of the trial. Cell purification. To maximize the safety of the anti-SSEA-1 antibody with regard to viral contaminations, we were initially committed to use an IgG-type of antibody and consequently ordered the development of such a customized clinical-grade product. Unfortunately, the antibody that was developed, although found to be ultimately free from any contamination, turned out to be unsuitable because of its poor affinity at the end of the purification process. We then had to redirect our attention towards a commercially available research-grade microbead-coupled IgM anti-SSEA-1 antibody (Miltenyi Biotec, Teterow, Germany) but were then requested by the regulatory agency to submit it to extensive viral testing (Supplemental Table 1) before it could be approved for use in our phase 1 clinical trial. Multiple preclinical runs were performed to optimize the sorting procedure which ended in a protocol that entailed the following steps : (1) withdrawal of the specification medium ; (2) wash-out of cells with phosphate-buffered saline -6- ([PBS] 10 mL/one B15 dish), detachment with trypsin (5 mL trypsin-EDTA [Hyclone] 0.25% per one B15; 4 minutes at 37°C) and blockade of trypsin by a buffer (PBS + Human Serum Albumin ([HSA, Albunorm, Octapharma, Boulogne-Billancourt, France] + 1% DNAse [Dornase alpha, Pulmozyme®, Roche, Boulogne-Billancourt, France]); (3) centrifugation of the cells (800 rpm, 4 minutes) which were then resuspended in the sorting buffer (PBS + HSA + 1% EDTA 2mM); (4) filtration of the cell suspension (100 µm) and cell counting in an hemocytometer; (5) additional cell washing in the sorting buffer before suspension in 100 µL of buffer for 10 million cells; (6) addition of the anti-SSEA-1 beads at a concentration of 20 µL per 10 million cells in 1 mL of the sorting buffer. Cells and beads were then incubated for 15 minutes at 4°C. They were then washed and resuspended in the same sorting buffer. The suspension was transferred into a LS column, up to 20 million cells per column, held within a magnet (Miltenyi Biotec). One column was used for 20 million cells. The magnetically labeled SSEA-1+ cells were eluted from the column after it had been removed from the magnetic field of the separator whereas the SSEA-1- cells were collected in the flow-through fraction. The positive fraction was again sorted on a second column, according to a similar procedure, to minimize contamination by still undifferentiated I6 cells and improve the purity of the cell population (≥95%). Specifications for raw and ancillary materials were sourced and documented as extensively as possible. Global gene expression analysis. RNA Extraction from ESC and ESC-derived CD15+ and CD15- cells: Total RNA was extracted from cells retrieved from three different batches using Trizol reagent (Roche). RNA integrity was evaluated by microfluidic analysis using the Agilent 2100 Bioanalyzer with an RNA LabChip® KIT (Agilent Technologie). RNA concentration was determined using a Nanovue (GE Healthcare). -7- Microarray Analysis : An aliquot of 200 ng of total RNA was used to synthesize doublestranded cDNA, and then produce biotin-tagged cRNA using the 3’IVT Express kit (Affymetrix®). The resulting bio-tagged cRNA were fragmented into strands of 35–200 bases in length according to the protocols from Affymetrix® (Santa Clara, CA, USA). The fragmented cRNA was hybridized to a Human Genome U219 array strip (Affymetrix®), containing more than 36,000 transcripts and variants. Hybridization was performed at 45°C in the Affymetrix® GeneAtlas™ Hybridization station for 20h. The GeneChip arrays strips were washed and then stained (streptavidin–phycoerythrin) on an Affymetrix® Fluidics Station 450, followed by scanning on the GeneAtlas™ Imaging Station (Affymetrix®). Normalization and Data Analysis : The hybridization data were analyzed using Expression Console (EC version 1.3.0.187, Affymetrix®). The scanned images were first assessed by visual inspection, then analyzed to generate raw data files saved as CEL files using the default setting of Expression Consol. Data were normalized using RMA algorithm. In a comparison analysis, genes were determined to be significantly differentially expressed with a selection threshold of ratio >2.0 or ratio <0.5 in the output result. Microbiological testing. The detailed list of the tests is indicated in the Supplemental Table 1. These tests were done by a subcontractor (Texcell, Evry, France) on pluripotent cells from both the MCB and late-production cell bank and were repeated on the SSEA-1-positive progenitor cells derived from these two banks. Viral testing was also performed on the anti-SSEA-1 antibody (hybridoma and final microbead-conjugated antibody used for cell sorting). Cytogenetic studies. Conventional cytogenetic analysis: Chromosome analyses by standard karyotype were performed from cultured cells using standard procedures (RHG and GTG bandings). For -8- mitotic preparations, cells were supplemented with 0.02 mg/ml colchicin (Eurobio, Courtaboeuf, France) for up to 1 h and 45 min. The cells were harvested and warm hypotonic solution of 0.075 M KCl was added in the preparation for up to 15-20 min. Finally, cells were fixed several times in cold Carnoy’s fixative (methanol/acetic acid, 3:1). Fluorescent in situ hybridization (FISH): FISH analyses were performed on interphasic nuclei and metaphase spreads. The centromeric probes specific for chromosome 12 and 17 were used according to the manufacturer’s recommendations (Vysis, Abbott France, Rungis, France). BAC clone RP5-857M17 specific for the 20q11.21 chromosomal region were used. Oligonucleotide based-array comparative genomic hybridization (array-CGH): The genomic imbalances were analysed by array-CGH using 105K oligonucleotide arrays (Hu-105A, Agilent Technologies, Massy, France). All array hybridizations were performed according to the manufacturer’s recommended protocols. In brief, 3 µg of genomic DNA was digested with AluI (5 units) and RsaI (5 units) for 2 h at 37°C and fluorescently labelled with the Agilent Genomic DNA labelling kit PLUS (Agilent Technologies). A male human genomic DNA (Promega, Charbonnière, France) was used as reference. Experiments were conducted in dyeswap. Cy5-dUTP patient DNA and its gender-matched reference labelled with Cy3-dUTP were denatured and preannealed with Cot-1 DNA and Agilent blocking reagent before hybridization for 40 h at 20 rpm in a 65°C rotating hybridization oven (Agilent Technologies). After washing, the slides were scanned on an Agilent Microarray Scanner. Captured images were processed with Feature Extraction 9.1 (Agilent Technologies) software and data analysis was performed with CGH Analytics 3.5 (Agilent Technologies). Copy number variations (CNVs) were considered significant if they were defined by three or more oligonucleotides spanning at least 50 Kb and contained at least one gene and were not identified in the Database of Genomic Variants. Fabrication of the fibrin patch. -9- Both fibrinogen and thrombin were components of the Evicel® kit (Ethicon Biosurgery, Omrix Biopharmaceuticals-Ethicon Biosurgery, Rhode Saint Genèse, Belgium) which is approved for clinical use and widely used as an hemostatic sealant in surgery. However, the fibrinogen/thrombin ratio had to be revisited as compared to what is indicated in the instructions for use of Evicel® to generate a scaffold meeting three major objectives : (1) an elasticity best suited for mesodermal gene expression (we ended up with an elasticity modulus of the patch, as measured by shear wave elastography, in the range of 6 kPa), (2) a fiber network dense enough to retain cells while remaining loose enough to prevent their excessively tight packing and to facilitate in vivo scaffold vascularization from the host vessels, and (3) a robustness compatible with easy intraoperative manipulations. At the end of the screening phase, concentrations of 20 mg/mL and 4 U/mL of fibrinogen and thrombin, respectively, were found to represent an appropriate combination, which is consistent with the finding that, at least with MSC, fibrinogen concentrations in the low range are best suited for promoting cell proliferation.3 SUPPLEMENTAL RESULTS Safety studies Among mice subcutaneously injected with cells mixed with matrigel, only those receiving undifferentiated hESC, SSEA-1-positive progenitor cells intentionally “contaminated” with 5% undifferentiated I6 cells and SSEA-1-negative cells prepared early in our first lab-scale experiments developed, although not consistently, a teratoma. Conversely, no teratoma developed in any of the 12 mice injected with 500,000 to 900,000 SSEA-1-positive progenitor cells (follow-up: 3 to 7 months), nor in those (n=8) exclusively injected with SSEA-1-negative cells generated at the completion of a selection step entailing the same protocol as the one - 10 - planned for the clinical trial (follow-up: 3 months). No teratoma was either seen in the group of mice (n=6) injected with SSEA-1-positive progenitor cells spiked with 2% undifferentiated I6 hESC. The 5 mice injected with a 10% fraction of undifferentiated cells mixed with the progenitors all died during follow-up, without apparent tumor but no necropsy could be made. Efficacy studies In keeping with data showing that hearts receiving the progenitor cell-loaded fibrin patch yielded the best recovery of LVEF, the cell/patch treatment was also associated with the greatest limitation of adverse LV remodeling, as demonstrated by a striking stability of LV enddiastolic volumes (mm3) : 361,74 [322.55;400.93] at 2 months vs 357.63 [323.89;391.37] at baseline. Conversely, these volumes markedly increased in hearts receiving a cell-free patch (380.79 [343.92;417.66] vs 365.12 [328.88;401.35] at baseline) and to a greater extent in the sham-operated rats (387.02 [341.59;432.45] vs 344.81 [319.38;370.23] at baseline; p = 0.034 vs the treated group). Changes in LV endsystolic volumes featured similar patterns as they increased (absolute difference between 2-month and baseline values, mean ± SEM) by 22.71 ± 12.56mm3 and 11.25 ± 13.85 mm3 in the sham and control groups, respectively, whereas they decreased by 14.92 ± 11.05 mm3 in hearts receiving the cell-loaded fibrin patch. SUPPLEMENTAL FIGURES LEGENDS Supplemental Table 1. Listing of the tests used for the viral testing. Supplemental Table 2. Summary of safety experiments in immunodeficient mice - 11 - Supplemental Figure 1. Morphological aspect of the undifferentiated I6 cell colonies throughout the scale-up process (P: Passage). Supplemental Figure 2. Expression of the cardiac transcription factors Isl-1 (panel A) and Mef2c (panel B) by the SSEA-1+ committed progenitor cell population embedded into the fibrin patch. For each panel, the top rows correspond to a patch loaded with undifferentiated I6 cells (negative control). Bar: 20 µm. Supplemental Figure 3. Expression of residual pluripotency (Panel A), mesodermal (Panel B) and endodermal (Panel C) genes in the CD15-negative fraction. These data were determined by a transcriptomic analysis based on 3 different sorting runs, using the AffymetrixGeneAtlasTM system coupled to the new Human Genome U219 array. Data are given as fold changes relative to undifferentiated I6 hESC. Blue bar graphs refer to the I6 line and red bar graphs refer to the CD15-negative fraction of the sorted cells. SUPPLEMENTAL REFERENCES 1. Amit M, Winkler ME, Menke S, Brüning E, Büscher K, Denner J, Haverich A, ItskovitzEldor J, Martin U. No evidence for infection of human embryonic stem cells by feeder cellderived murine leukemia viruses. Stem Cells. 2005;23:761–771. 2. Amit M, Itskovitz-Eldor J. Derivation and spontaneous differentiation of human embryonic stem cells. J Anat. 2002;200:225–232. 3. Ho W, Tawil B, Dunn JCY, Wu BM. The behavior of human mesenchymal stem cells in 3D fibrin clots: dependence on fibrinogen concentration and clot structure. Tissue Eng. 2006;12:1587–1595.