Template for Electronic Submission to ACS Journals
advertisement

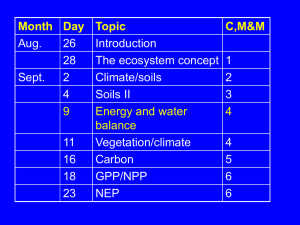
The binding of quinone to the photosynthetic reaction centers. Kinetics and thermodynamics of reactions occurring at the QB-site in zwitterionic and anionic liposomes. Supplementary material (A) Table S1. Start guess kinetic constants for PC and PG liposomes at 25°C (see Figure 1); is obtained using KQWAM . kout Kinetic Constant PC start guess Δk/k % PG start guess Δk/k % (s−1) kout 154 3 44 3 kBA (s−1) 487 0.4 97 0.3 kout (s−1) 500 17 500 16 RMSD 1.49% 1.60% R2 0.992 0.992 1 (B) Figure S1. Experimental decays (black lines) of D+ for PC (upper panel) and PG (lower panel) at 25°C contrasted with the ODE set calculated data (grey lines) 2 In this figure the experimental traces at 25°C for PC and PG and the best fit curves obtained from ODE set calculation are shown. Considering that all the traces at the different Q/RC ratios are simulated simultaneously and that the optimization is performed with only two parameters, the agreement with the experimental data is satisfactory. 3 * (C) Figure S2. R2 as function of optimized kout for PC and PG at the temperatures indicated in the legend. A set of independent optimizations were performed using twenty values between 1 and 100 s−1 as fixed values for kout . For each run, new optimized values of k*in and kin are obtained. The 4 dependence of the R2 values on kout is shown for PC and PG. By varying kout in the above mentioned range, the exchange regime is forced to change from slow to fast and the simulated decays correspondingly show a different agreement with the experimental CRR traces. The shape of the obtained curves is similar at all temperatures: R2 is lower at low kout values, then increases and reaches a plateau for higher values, indicating that the fast quinone exchange is always a good mathematical description of the system. However, the value of kout at which the R2 value starts to decrease can be taken as the kout lower limit. Table S2. Estimated upper (having set kin at its higher value) and lower (from figure S2) limits of kout in PC and PG liposomes t(°C) (s–1) kout lower PC (s–1) kout upper PC (s–1) kout lower PG (s–1) kout upper PG 6.5 20 35 3 3.5 10 20 33 3 4.5 15 20 53 5 10 20 20 80 8 15 25 20 146 8 36 30 20 238 8 72 35 20 450 8 150 5 (D) Correlation among kinetic parameters. We studied the Hessian matrix of the sum of quadratic deviations between experimental data and ODE set numerical solutions (χ2) as a function of the optimized physical parameters of our model. This study provides different and useful kinds of information about the actual dependency of the goodness of the fit upon the parameters and therefore about their relevancy to the model. However, the parameter variations will most probably result coupled by the set of equations describing the model and the coupling coefficients will often have a physical significance and provide additional insight into the working of the model. To this effect, the Hessian matrix is diagonalized. True minimization respect to the Hessian variables is ensured by the emergency of all positive eigenvalues. The smallest eigenvalues are those to be watched upon with more attention since they signify the possibility of greater deviance from the minimum values within the same confidence interval when moving along the associated eigenvector. Since the value of parameters fixed along the optimization was the result of independent experiments, their confidence should be separately assessed. However the impact of variation of these parameters upon the results of optimization is of obvious interest as well. We therefore evaluated how RMSD is affected by the above said variation. Both the gradient with respect to the fixed parameters and the Hessian matrix with respect to the optimized ones were obtained by means of finite difference using a displacement of 1% from the reference value. The following formulas were used for evaluating the gradient components: 2 (kopt k ) - 2 (kopt ) 2 kopt (S2.1) ln k k 6 and the Hessian matrix: a a2 2a0 2 2 2 1 kopt (S2.2) 2 2 ln k kopt a a00 a01 a10 2 2 11 ka kb (S2.3) ln ka ln kb ka kb where a0 = χ2(kopt), a1 = χ2(kopt + Δk), a2 = χ2(kopt - Δk) and a00 = χ2(ka, opt, kb, opt), a10 = χ2(ka, opt + Δka, kb, opt), a01 = χ2(ka, opt, kb, opt + Δkb), and a11 = χ2(ka, opt, + Δka, kb, opt + Δkb). Since logarithmic derivatives are used, gradient and Hessian are describing χ2 change against relative variation of the parameters. The obtained matrix is diagonalized and the eigenvalues are extracted. The Hessian matrix represents a parabolic approximation of the χ2 dependence on the deviation of parameters from their optimal values. The eigenvalues D are associated to the convexity of the χ2 paraboloid along the V column vector; a high D value indicates that χ2 grows faster along the associated V direction and that the parameter V, linear combination of the original parameters, is obtained with good accuracy, P(χ2)/√𝐷 being the actual probability distribution for deviation along the direction V. To ease the interpretation of the data we represent here only 2x2 sections of the complete hessian matrix. As already mentioned, k*in and k*out are expected to be strongly correlated since their ratio gives K*Q. This is mathematically demonstrated by the eigenvalues (D) and the eigenvectors (V) of the Hessian. 7 Table S3.1 Correlation table for kin and kout V1 V2 kin -0.714 -0.700 kout -0.700 0.714 √𝐷 0.04 0.5 % err 12 1.0 Table S3.1 shows the hessian analysis results for kin and kout and evidences that the two parameters are strongly correlated. kin and kout can move in the same direction (V1), leaving K Q unaltered, without significant χ2 change; on the other side vector V2 representing the fastest direction for K Q change has a large associated D value. This shows that K Q is a meaningful parameter for the present set of experiments and that it can be obtained with a much better precision (about 10 times) than the individual dividends. On the other hand, the contributions of LAB and K Q can be considered well separated as demonstrated by the relevant V and D, calculated on the basis of kout and kBA, shown in Table Sb. Table S3.2 Correlation table for kout and kBA. V1 V2 kout -0.977 0.213 kBA 0.213 0.977 √𝐷 0.25 1.1 8 1.8 % err 0.4 The V eigenvectors are approximately directed along the parameter directions and are in both cases determined within a fair error. This finding is important since in previous works done in detergent it was not possible to obtain reliable thermodynamic parameters for the long chain quinone binding constant to the QB-site due to the temperature dependence of the micellar properties.1 The other parameters analyzed for correlation are kout and kout: Table S3.3 Correlation table for kout and kout. V1 V2 kout -0.184 -0.983 kout -0.983 0.184 0.028 0.35 17 1.3 √𝐷 % err and kout are only weakly correlated since V1 and V2 are directed along the main axes. As a kout consequence, the values of K Q and KQ are completely independent, K Q being well determined and KQ being less relevant for the model. 1. McComb, J. C.; Stein, R. R.; Wraight, C. A. Investigations on the influence of headgroup substitution and isoprene side-chain length in the function of primary and secondary quinones of bacterial reaction centers. Biochim Biophys Acta 1990, 1015 (1), 156-71. 9 10