tpj12187-sup-0010
advertisement

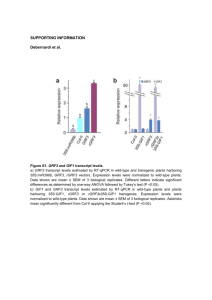
Supplemental figure legends Figure S1. Multiple sequence alignment of CSP amino acid sequences. Four Arabidopsis CSPs (deduced from AtCSPs1-4: At4g36020, At4g38680, At2g17870, At2g21060, respectively), Escherichia coli CspA (AP_004238.1) and Homo sapiens YB-1 (NP_004550.2) were aligned by CLUSTAL_W. Gaps were introduced to ensure maximum homology. Identical amino acids are indicated by an asterisk and semi-conserved substitutions are indicated with a colon. Yellow highlight indicates the region used to synthesis a CSP1-specific peptide for polyclonal antibody production. Red highlight indicates a conserved Serine phosphorylation site found in YB-1 (Ser102) (Evdokimova et al., 2006). Black underline indicates the conserved CSD domain. Figure S2. Cold increases CSP1 accumulation. (a) Plant growth, treatment and harvest scheme. (b) Three-wk-old plants (35S:HF-RPL18) were maintained at 23ºC or transferred to 4ºC at ZT2 and harvested at the indicated times over a 48 h period. SDS-solubilized proteins of rosette leaves were separated by 12.5% (w/v) SDS-PAGE, and subjected to immunoblot analysis to detect CSP1 and FLAG-RPL18 using an antiserum prepared against a specific peptide of CSP1 and anti-FLAG antiserum, respectively. The apparent molecular mass of CSP1 is 30 kDa and FLAGRPL18 is 24 kDa. Figure S3. atcsp1-1 is a knock-down allele. (a) Diagram showing the position of the T-DNA insertion and primers (arrows, 1:AtCSP1geneF, 2: LBa1, 3: AtCSP1geneR) used for genotyping. (b) Total RNA from Col-0 and atcsp1-1 homozygotes was extracted from 10-d-old seedlings and semi-quantitative RT-PCR was performed and separated on agarose gels. ACTIN7 expression was used as a loading control. (c) CSP1 transcript abundance in leaves of 25-d-old plants maintained at 23ºC or transferred to 4ºC for 12 h. Data are total mRNA measured in the microarray analysis relative to Col-0 (Table S1b). Figure S4. Cold increases levels of CSP1 in polysomes. Polysome absorbance profiles from 14d-old seedlings of three genotypes, atcsp1-1 (a), 35S:HF-RPL18 (b) and 35S:AtCSP1-FH#11 (c), grown under non-stress conditions (NS: black line) or exposed to 4oC for 9 h (Cold: red line). Proteins from each sucrose gradient fraction were precipitated, fractionated by 12.5% (w/v) SDSPAGE, and subjected to immunoblot analysis with anti-CSP1 (CSP1) or anti-RPS6 (S6) antisera. The anti-CSP1 antiserum detects both endogenous CSP1 and CSP1-FLAG of 35S:AtCSP1-FH seedlings. The apparent molecular mass of CSP1, CSP1-FLAG and RPS6 are 30, 34 and 28 kDa respectively. Figure 3b is shown here as panel b to facilitate comparison of genotypes. (d) Table summarizing developmental age and low temperature conditions tested in polysome profile analyses. In all cases, no significant change in polysome levels was observed in response to cold stress. Figure S5. Co-immunopurification of RNA with CSP1 is not due to non-specific binding and purification. Whole rosette tissue of 35S:AtCSP1-FH#11 following cold treatment (4ºC for 12 h) was used for IP with -CSP1 coupled to Protein G beads and anti-HA mAb coupled to Protein G beads, as a negative control. Comparative capillary electrophoretic analysis of CSP1 IP RNA and -HA mAb IP RNA. CSP1 IP yielded approximately 1 ng RNA per mg frozen pulverized rosette leaf tissue and HA mAB IP yielded less than 0.01 ng RNA per mg tissue. Figure S6. Validation of CSP1-target enrichment by immunopurification. (a) qRT-PCR analysis of selected transcripts using RNA samples obtained from total and CSP1-IP (IP) mRNAs of whole rosettes of 35S:AtCSP1-FH#11 (OE) or 35S:HF-RPL18B (WT) plants grown at 23ºC (NS) or transferred to 4ºC (Cold) for 12 h. Values are relative to ADK2. (b) Means of microarray MAS5.0 signal values from total and CSP1-IP mRNA. Values in parentheses are the percent of detected signal value compared to those derived from total mRNAs of each genotype under NS condition. Figure S7. Immunoblot analysis of CSP1 levels in genotypes used in this study. Fifteen μg protein extracted from 3-wk-old rosettes of two independent overexpression lines (35S:AtCSP1FH#4, 35S:AtCSP1-FH#11), Col-0, and atcsp1-1, were separated on a 12.5% (w/v) SDS-PAGE and transferred onto a nitrocellulose membrane. Immunoblot detection was performed with antiCSP1 or anti-RPS6 antisera as described for Figure 1h. Table S1. DNA microarray analysis of total, polysomal and CSP1-immunopurified mRNAs under non-stress and cold-treatment conditions. Table S2. Primers used for gene constructs and quantitative RT-PCR.