Amplification and re-generation of LNA
Pending illustrations/experiments
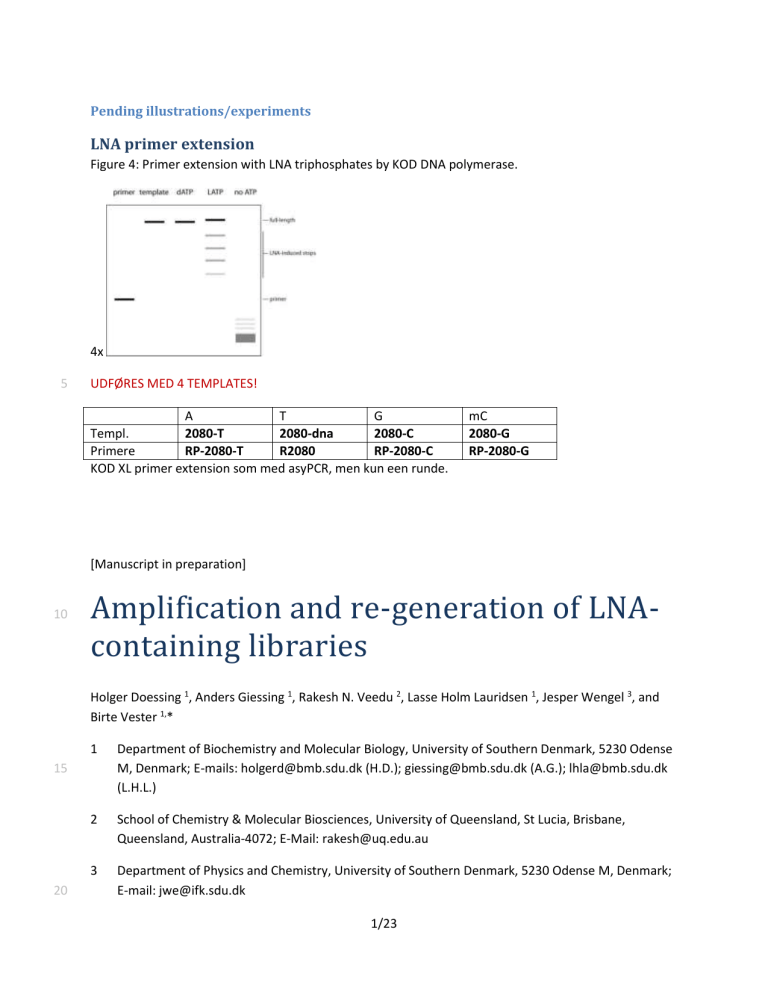
LNA primer extension
Figure 4: Primer extension with LNA triphosphates by KOD DNA polymerase.
5
10
4x
UDFØRES MED 4 TEMPLATES!
Templ.
A
2080-T
T
2080-dna
G
2080-C
Primere RP-2080-T R2080 RP-2080-C
KOD XL primer extension som med asyPCR, men kun een runde. mC
2080-G
RP-2080-G
[Manuscript in preparation]
Amplification and re-generation of LNAcontaining libraries
15
20
Holger Doessing 1 , Anders Giessing 1 , Rakesh N. Veedu 2 , Lasse Holm Lauridsen 1 , Jesper Wengel 3 , and
Birte Vester 1, *
1 Department of Biochemistry and Molecular Biology, University of Southern Denmark, 5230 Odense
M, Denmark; E-mails: holgerd@bmb.sdu.dk (H.D.); giessing@bmb.sdu.dk (A.G.); lhla@bmb.sdu.dk
(L.H.L.)
2 School of Chemistry & Molecular Biosciences, University of Queensland, St Lucia, Brisbane,
Queensland, Australia-4072; E-Mail: rakesh@uq.edu.au
3 Department of Physics and Chemistry, University of Southern Denmark, 5230 Odense M, Denmark;
E-mail: jwe@ifk.sdu.dk
1/23
* Author to whom correspondence should be addressed; E-mail: b.vester@bmb.sdu.dk; Tel.: +45-
6550-2406; Fax: +45-6550-2467.
2/23
25
Abstract
*ca. 200 ord
Keywords
Aptamer, in vitro selection, locked nucleic acid (LNA)
3/23
30
35
Introduction
*BIRTES FORSLAG TIL OPBYGNING: Vi *har* forsøgt selektioner med de ’tidligere publicerede’ protokol
(med små ændringer), men det virker sgu’ ikke!?! Derfor vil vi nu gerne belyse *hvorfor*! Er det fordi vi mister vores LNAer? (med diversitetstab som konsekvens heraf)*
*KONKLUSION: Hermed har vi fået syn for, at LNA-indholdet falder en smule, men er det kritisk?
Derudover har vi fået stablet nogle nye metoder på bordet til fx at undersøge LNA-indholdet i biblioteker*
*LNA er dejligt*
The first LNA triphospate to be synthesized was LNA adenosine triphosphate (LNA ATP), soon followed by LNA TTP, LNA 5-methyl-CTP, and LNA GTP. *REF*
This opened up the field of enzymatic applications, and we [1-5] and others [6] have previously
established how Phusion DNA polymerase and KOD DNA polymerase are both able to use an LNAmodified DNA template as well as incorporate LNA into DNA strands
Figure 1. Structures of locked nucleic acid triphosphates LNA ATP and LNA TTP.
LNA is characterized by the 2 ′ -4 ′ bridge that locks the ribose into the 3 ′ -endo conformation normally observed in A-form duplexes.
40
45
LNA ATP LNA TTP
*ALLE DE FORSKELLIGE KOMBOS RAKESH LAVEDE MED POLYMERASER OG TEMPLATES/PRODUKTER*.
The aforementioned properties of LNA meant that
*Vi har tidligere vist, at vi kan indbygge LNA vha. polymerase*
*SELEX er løsningen på alt*
*Vi tror, vores setup kan bruges til SELEX*
Here we introduce a scheme for amplification and re-generation of a randomized pool of LNA-containing oligomers. We successfully incorporated either LNA T or LNA A in two DNA libraries and found that after
4/23
three rounds of amplification and re-generation the LNA content in our pools remained at least 8%, suggesting that the scheme is feasible for in vitro selection of LNA-containing oligonucleotides.
5/23
50
55
60
65
Results
Phusion DNA polymerase reads LNA-containing strands
Library design
We wanted to find out whether we could employ the previously established LNA-compatible polymerases, KOD DNA polymerase and Phusion DNA polymerase, to successfully amplify and regenerate a library of LNA-containing single-stranded sequences. We based our library design on the following guidelines: (1) No LNA moieties in the primer binding regions; (2) *****
Phusion DNA polymerase can employ LNA templates
Phusion DNA polymerase had previously been shown to be able to amplify an LNA-containing template in a PCR setup, *REFS* however, the templates employed were fairly short (*XXX* nucleotides) and their LNA content and distribution did not correspond well with a typical in vitro selection library. We therefore asked whether Phusion DNA polymerase could indeed amplify a ‘long’ (>60 nucleotides) LNAcontaining template.
We prepared the LNA A-containing template shown in Figure 2A and found that Phusion DNA
polymerase was able to successfully generate a double-stranded all-DNA product of the expected size
deoxy-ATP we were unable to obtain the desired product (lane 2). We were unable to find any experimental conditions that would generate a suitable double-stranded LNA-containing product (data not shown), indicating that Phusion DNA polymerase is not fit for synthesis of double-stranded LNAcontaining products of this size and composition.
Figure 2: PCR amplification with LNA ATP by Phusion DNA polymerase.
(A) Sequence of the template employed. The library ‘core’ is highlighted in blue. Lowercase ‘a’ is LNA A.
Primers and their binding sites are indicated. (B) Agarose gel electrophoresis of PCR products. Lane 1:
PCR with all four deoxynucleotide triphosphates. Lane 2: PCR, dATP substituted by LNA ATP. Lane 3:
Control PCR without ATP. Lane 4: Control PCR without template.
A
5 ′ -GGTCTGGTCCACACCCAG
5 ′ -GGTCTGGTCCACACCCAG CCGCCaCCCaGGGaCGCaGCCaGGCaCGGCG GGCCTATAGTGAGTCGTATTA-3 ′
3 ′ -CCGGATATCACTCAGCATAAT-5 ′
6/23
B
70
75
80
Phusion DNA polymerase correctly reads LNA
The fidelity of Phusion DNA polymerase reading an LNA A-containing template was tested using an LNA containing library with a core of 25 randomized positions (C, G or T) and 7 LNA A-moieties at fixed
selection library, and (b) it allowed us to quickly identify LNA A-related sequence errors in the product.
The library was amplified by PCR with deoxyribonucleotide triphosphates, and the corresponding double-stranded DNA was then cloned into a bacterial plasmid vector and sequenced. *REF TIL
MINIARTIKLEN*
Figure 3B shows a sequence logo obtained from alignment of the 32 positively identified member
sequences. All LNA A positions were clearly identified in all members. Three members contained singlenucleotide insertions and one member had a single-nucleotide deletion, but neither of these involved adenosines. Whether these minor errors stem from our library preparation, the PCR or even the vector’s maintenance in the bacterial host is unclear. We conclude that Phusion DNA polymerase correctly reads
LNA A moieties that are spaced three or more nucleotides apart in a DNA library.
Figure 3: Phusion DNA polymerase correctly reads an LNA A-containing DNA template.
(A) Sequence of the DNA library used. Lowercase ‘a’ denotes LNA A moieties; ‘B’ denotes C, G or T. (B)
Sequence logo of the randomized core of the sequenced library members (n = 32). Letter height indicates relative abundance. All LNA A-positions were clearly identifiable in all 32 members. Positions
1, 20 and 30 are alignment artifacts caused by single-nucleotide insertions in three members. Another member had a single-nucleotide deletion (aligned to pos. 25).
A
5 ′ -GGACAGGACCACACCCAG-aBBBBaBBBaBBBaBBBaBBBaBBBaBBBB-GGCCTTTTGTGTGTCGTTT-3 ′
7/23
B
85
90
KOD DNA polymerase can incorporate LNA triphosphates
KOD DNA polymerase accepts all four LNA triphosphates
KOD DNA polymerase was previously shown to be able to incorporate LNA *XXX* triphosphates under primer extension conditions. *REF* We extended this analysis by attempting primer extension with three deoxy-triphosphates plus either of the four LNA triphosphates on *XXX* (*FIG*).
* primer ext med alle fire LNA-TPer *
Figure 4: Primer extension with LNA triphosphates by KOD DNA polymerase.
(A) Template sequences for primer extension experiments where one LNA is substituted for the corresponding DNA triphosphate. Template sequence is black, primer is blue. Lowercase indicates LNA moieties in product (grey).
(B) Denaturing polyacrylamide gel electrophoresis (PAGE) of primer extension reactions with ***
8/23
A
**HOV?!! DER BLIR JO LNA-INKORPORERING I PRIMER-REGIONERNE
MED DETTE SETUP!!?!
Primer extension with LNA ATP
5 ′ -GGACAGGACCACACCCAG VVVVVVVTVVVVVVTVVVVVVTVVVVVVTVVVVVV GGCCAAAAGAGAGACGAAA-3 ′
3 ′ -CCTGTCCTGGTGTGGGTC BBBBBBBaBBBBBBaBBBBBBaBBBBBBaBBBBBB CCGGTTTTCTCTCTGCTTT -5 ′
Primer extension with LNA TTP
5 ′ -GGACAGGACCACACCCAG ABBBBABBBABBBABBBABBBABBBABBBB GGCCTTTTGTGTGTCGTTT-3 ′
3 ′ CCtGtCCtGGtGtGGGtC tVVVVtVVVtVVVtVVVtVVVtVVVtVVVV CCGGAAAACACACAGCAAA -5 ′
Primer extension with LNA GTP
5 ′ -GGACAGGACCACACCCAG DDDDDDDCDDDDDDCDDDDDDCDDDDDDCDDDDDD GGAAGGTTGTGTGTAGTTG-3 ′
3 ′ CCTgTCCTggTgTgggTC HHHHHHHgHHHHHHgHHHHHHgHHHHHHgHHHHHH CCTTCCAACACACATCAAC -5 ′
Primer extension with LNA m5-CTP
5 ′ -GGACAGGACCACACCCAG HHHHHHHGHHHHHHGHHHHHHGHHHHHHGHHHHHH CACCTTCCATACATCATCC-3 ′
3 ′ ccTGTccTGGTGTGGGTc DDDDDDDcDDDDDDcDDDDDDcDDDDDDcDDDDDD GTGGAAGGTATGTAGTAGG -5 ′
B
9/23
95
100
105
110
115
120
125
An amplification scheme for LNA strands
Template preparation with lambda exonuclease
We have previously described a strategy for propagating a DNA library with multiple LNA modifications at defined positions using Phusion High Fidelity DNA polymerase for amplification and KOD DNA
polymerase for regeneration [7]. Here, we extend this protocol to include propagation of a fully
randomized DNA library where either adenosine or thymidine has been replaced by their corresponding
LNA nucleosides. We use KOD XL DNA polymerase rather than regular KOD DNA polymerase, as the XL
(data not shown *ELLER LASSES DATA I SUPPLEMENTARY?*).
Carry-over of the DNA template from one selection round to the next is generally undesirable; it dilutes the selected sequence pool by re-introducing noise in the form of unspecific members. In vitro transcribed libraries are often treated with DNase I in order to remove the DNA templates. This approach cannot be used with pools of single-stranded DNA, where other methods are used: (1) gel shift followed by purification from acrylamide gels; (2) *FLERE?!*.
Since we wanted to confine LNA to the randomized region we could not simply digest the DNA with an exonuclease, as this would also have affected the primer binding sites of our pool. Instead, we opted for a two-part strategy: First, removal of the non-template strand with the strongly processive lambda
exonuclease, which specifically digests the phosphorylated strand in double-stranded DNA [9].*REFS*
Second, after LNA strand synthesis full-length oligomers were extracted by oligo capture.
PCR amplification of the LNA-containing pool was with a 5 ′ -phosphorylated forward primer and a fluorophore-labeled reverse primer. Complete digestion by lambda exonuclease was observed as the inability of the single-stranded digestion products to bind ethidium bromide as well as a slight gel shift of the fluorescently labeled template strand (**Ошибка! Источник ссылки не найден.).
Asymmetric PCR increases yield over primer extension
Primer extension could be extended to what is effectively an asymmetric PCR with KOD DNA polymerase. This greatly increased our yield of full-length LNA-containing strands. We found that after
approach could also be used with the KOD XL DNA polymerase formulation. *EVT REF TIL LASSES SML.
TEST MED KOD OG KODXL I SUPPLEMENTARY*
Purification of full-length LNA strands
* dynabeads *
LNA is retained over multiple rounds
LNA is the natural substrate for neither Phusion nor KOD XL DNA polymerase and it is conceivable that library members with little or no LNA content would be preferentially amplified in our selection scheme.
We therefore sought to determine the impact our protocol had on the LNA content in our pool after a number of rounds of amplification and regeneration. Importantly, we performed our rounds without
10/23
130
135
140 selection against a target ligand. The only selection pressure on our library was therefore imposed by the amplification procedure itself.
1.
PCR with Phusion DNA polymerase and DNA triphosphates to obtain the corresponding doublestranded DNA. The primer for the non-template strand carried a 5 ′ phosphate.
2.
Purification of the DNA on a spin column.
3.
Digestion of the phosphorylated strand by lambda exonuclease yielding the single-stranded template DNA.
4.
15 rounds of asymmetric PCR with KOD XL DNA polymerase and an LNA triphosphate and the additional three DNA triphosphates.
5.
Isolation of the full-length LNA strands by oligonucleotide capture on magnetic beads.
6.
Careful washing of the LNA strands, followed by heat-elution into 1×SSC. A fraction of the eluted
LNA strands were then used as template in step 1 for another round of amplification.
We performed 3 such amplification rounds using either LNA ATP or LNA TTP. (For the latter the library’s complementary strand acted as template.)
Figure 5: Schematic of our protocol for amplification, regeneration of an LNA-containing library.
DNA is blue, LNA is red. Starting from a DNA library (1), we introduce a fluorophore and phosphate by
PCR (2). The phosphorylated strand is then enzymatically removed (3) to yield the single-stranded template for primer extension with LNA and DNA triphosphates (4). Full-length strands can be captured with a streptavidin bead-bound dual biotinylated probe complementary to the products’ 3′ end (5), and, after washing, results in a clean eluate (6). In vitro selection proceeds with ligand binding and partitioning into binders and non-binders (7). When the library has achieved sufficient binding affinity it is cloned into a vector and sequenced for further analysis (8).
Here, we bypass step 7. Instead, we analyze the composition of the LNA eluates from 3 consecutive cycles directly by an exonuclease assay and indirectly by LC/MS analysis of the PCR products. We also perform sequencing analysis of the initial library and eluates from the third cycle (red arrows).
11/23
145
150
155
The pools contain LNA after 3 rounds
The LNA content of the LNA strands obtained from rounds 1-3 were subjected to nucleolytic digestion by KOD DNA polymerase. Unlike the KOD XL variant this polymerase has a very aggressive 3-5 ′
(our unpublished data). Incubation of the LNA strands with KOD DNA polymerase yielded only partially
digested products (Figure 6C/D, lanes 1-6), whereas the crude DNA library was fully digested (Figure
First, digestion stops – visible as a smear – are confined to a region corresponding to the LNA-encoding
rates in accordance with a uniform distribution of LNA moieties within the randomized cores.
12/23
160
The exponential decay rates (slopes) decrease slightly from round 1 to round 3. This indicates that fewer
LNA moieties are present near the 3 ′ end of the LNA strands rounds 2 and 3 and so the exonuclease can digest further into the random region. This effect is most pronounced with LNA A-containing strands
nucleolytic attack than LNA T-containing strands, as reflected in the lower decay constants; this could be due to a higher LNA content and/or higher nuclease stability of the LNA T-containing strands (compare
red curves in Figure 6E and Figure 6F).
Figure 6: LNA content of libraries maintained for 3 rounds with either LNA ATP or LNA TTP.
(A) An N40 library (40 fully randomized nucleotides) was used as template. Extension of the top primer yields a pool with adenosines in the N
40
region only; extension of the bottom primer yields a pool with thymidines in the N
40
region only. (B) Our amplification/regeneration scheme for making
LNA-containing strands. We did 3 rounds of amplification/regeneration with either LNA ATP or LNA
TTP in order to test whether our amplification scheme entails intrinsic selection pressure against LNAcontaining pool members. LNA content of strands from round 1-3 was assayed by digesting with a 3 ′ -
5 ′ deoxyribonuclease (KOD DNA polymerase) that cannot digest past LNA moieties. (C, D) Digestion analysis of strands amplified using LNA ATP (C) or LNA TTP (D). Lanes 1-6 show 5 ′ -radiolabeled LNA strands incubated at 37 °C/20 min. without or with nuclease. The position of the LNA-induced digestion stops is consistent with the expected position of the LNA moieties in the oligomer pools.
Lanes 7, 8: The all-DNA crude library is fully degraded. Lanes 9, 10: Digestion of a chemically synthesized LNA A-containing oligomer yields stops at the expected intervals. The digestive patterns shown here all remained virtually identical after 80 min. (not shown). (E, F) Densitometry scans of lanes 1-6 in A and B. Total pixel intensities of lane pairs (i.e. with and without nuclease) were matched, and all pairs were then normalized for direct comparison. Blue curves indicate ‘without nuclease’, red curves indicate ‘with nuclease’. Curve brightness reflects round number. (E) As expected for a fully randomized library the intensity of the nuclease stops fits an exponential decay function. The decay slope decreases from round 1 to round 3, indicating that the LNA A content in the pool decreases. (F) Similarly, the LNA content also decreases in a pool propagated with LNA TTP.
However, the densitometry curves are higher and steeper than for the LNA ATP-propagated pool, suggesting that LNA TTP offers better incorporation and/or higher stability.
A
*Første del af figuren burde måske være noget med at vise amplifikations-setuppet.*
B
Extend with LNA ATP →
5 ′ -GCTCTGTCGTCGTGGTCGGTCC-3 ′ 0 /GCCTTTTGTGTGTCGTTT-3
′
5 ′ -GCTCTGTCGTCGTGGTCGGTCC N
40
GGCCTTTTGTGTGTCGTTT-3 ′
3 ′ -CGAGACAGCAGCACCAGCCAGG N
40
CCGGAAAACACACAGCAAA-5 ′
3
′
-CGAGACAGCAGCACCAGCCAGG N 3 ′CCGGAAAACACACAGCAAA-5 ′
← Extend with LNA TTP
13/23
C D
E F
165
170
Successive regeneration cycles cause a small decrease in LNA content
Next, we wanted a quantitative measure of the LNA content in our pool. We approached this by analyzing the nucleoside composition of our LNA strand pools. This can be done by digesting the DNA of
interest with nuclease P1, phosphodiesterase I, and alkaline phosphatase [10] and subjecting the
resulting nucleoside mix to liquid chromatography-mass spectrometry (LC/MS). However, due to the nuclease resistance of (successive) LNA moieties we opted to obtain the corresponding double-stranded
DNA by PCR. By using a dual-biotinylated primer for the template-encoding strand we were able to immobilize the DNA on streptavidin-coated magnetic beads and subsequently heat-elute the complementary strand. The eluted single-stranded DNA oligomers therefore contained the same sequence content as the original LNA strands and could easily be quantitatively hydrolyzed to their nucleoside constituents and analyzed by LC/MS.
14/23
175
180
with LNA ATP decreased from 12% to 8% over three rounds. Similarly, thymidine content in DNA
fit with our nuclease digestion assay, which also indicated a decrease in LNA content from round 1 to
The specific decrease in the number of LNA-corresponding nucleosides indicates that our amplification/regeneration setup does impose a certain level of selection pressure against LNAcontaining strands. We note, however, that in neither case did we find a sudden decrease in LNA content from one round to the next.
Figure 7: Nucleotide composition of libraries maintained for 3 rounds with either LNA ATP or
LNA TTP.
The N40 DNA library was amplified and regenerated for 3 rounds with either (A) LNA ATP or
(B) LNA TTP. The nucleotide composition of the libraries was determined by isolating one strand of the corresponding PCR product, digesting it to deoxyribo-nucleosides, and quantitating the mixture by LC/MS. The nucleoside corresponding to the LNA species is highlighted in bold. Round 0 corresponds to the crude DNA library.
A B
Library maintained with LNA ATP: Library maintained with LNA TTP:
12% 11% 9% 8%
33% 36%
44%
39%
30%
29%
33%
27%
12% 10%
8%
30%
33%
7%
28%
31% dT dA dG dC
25%
37%
32%
36% dT dA dG dC
25% 22% 21% 22%
30%
25%
29%
28%
185
190
0 1 2 3
Rounds of amplification, regeneration
0 1 2 3
Rounds of amplification, regeneration
Sequence analysis indicates fewer ‘LNA islands’
Although Phusion High Fidelity DNA polymerase can read an LNA-modified DNA template with good fidelity it is conceivable that the enzyme might stall at sites containing successive LNA monomers.
Likewise, it is possible that the rate of nucleotide incorporation by KOD XL DNA polymerase might be very low at sites requiring multiple incorporations of LNA triphosphate. In this is the case, we might expect the number of e.g. LNA di-, tri-, and tetra-meric (or higher) LNA sites in a library to be reduced
15/23
195
200
205
210 over consecutive rounds of amplification and regeneration. This would significantly decrease the diversity of the resulting LNA-containing pool. To find out if this was the case, we looked at the sequence composition at the basal level by sequencing. Clones were obtained by amplifying either the crude DNA library or LNA A- or LNA T-containing pools from round 3 with Phusion DNA polymerase and cloning the resulting DNA into a plasmid vector. Sequencing yielded *XXX FOR URU0*, *XXX FOR
URU3A*, and *XXX FOR URU3T* clones, respectively.
We then analyzed the sequence data and counted how many occurrences of A
4
, A
3
, A
2
, and singular adenosines (flanked by non-adenosines) each of the three libraries encoded. (Note that since the LNA Tmaintained library was simply prepared using the reverse complement of the crude library, any LNA thymidine in the pool is simply represented by an adenosine in the presented sequence data. *ELLER
OGSÅ SKAL JEG BARE SKILLE FIGUREN AD I TO*) From this we could calculate the frequency of each of
these motifs. The results are shown in Figure 8.
*Neither* of the libraries contained the A
4
motif. While the crude library encoded the A
3
motif at a frequency of *0.2* this motif was not found in libraries maintained for 3 rounds with LNA ATP or LNA
TTP. Similarly, while the crude library contained the A
2
motif at a frequency of *1.3 * per member, much fewer members of the LNA-maintained libraries contained this motif, ***
The average distance *BEMÆRK: AFSTAND OG GENNEMSNITLIGT ANTAL ER _IKKE_ DET SAMME!! men begge tal er måske interessante at tage med?* between adenosines within the random region in the crude library was *N.N±N.N* nucleotides. This distance decreased to *X.X±X.X* nucleotides for the LNA
A-containing pool and *Y.Y±Y.Y* nucleotides for the LNA T-containing pool.
Notably, we did not see similar marked changes in the distribution of cytidines, guanosines, and
‘locked’ nature and consequent lesser compatibility with either Phusion High Fidelity DNA polymerase or
KOD XL DNA polymerase, or both.
Figure 8: Comparison of nucleotide distributions in the crude library and libraries maintained for 3 rounds with LNA ATP or TTP.
**
Left: Column height reflects the frequency with which the indicated number of successive adenosines (flanked by non-adenosines) occurs within a library member. Frequencies are also indicated above each column.
Right: Similar plots indicating the frequencies of successive cytidines, guanosines, and thymidines, respectively.
*NB: beware that commas are not periods!*
*NOTE: LNA T-biblioteket undersøges for A-indhold – fordi vi kigger på den revcompl. streng!*
16/23
215
6
5
4
3
2
1
0
05
Crude DNA library
LNA A, 3 rounds
LNA T, 3 rounds
4
01
01
00 00
3 2
No. of successive adenosines
03
03
**INDSÆT GTC
HER*
1
17/23
220
225
230
235
Discussion
Given that typical in vitro selection experiments are concluded within *X-XX* rounds *REFS*, we do not expect that this methods’ intrinsic selection pressure is significantly strong to entirely eliminate LNA from the pool, however. After 3 rounds, LNA content is 8%; this corresponds to approximately 6 LNA moieties per molecule (i.e. 15% LNA in the randomized core). Provided that the conditions of an in vitro selection experiment do favor nucleic acid structures with e.g. stabilized stem-loops even a single LNA moiety could still have a substantial impact on selection outcome. Moreover, if further LNA moieties are desired in the final pool the randomized can may be lengthened and/or spiked with further LNAencoding positions. It should also be noted that we started out from a DNA library; if in vitro selections are initiated from an LNA-containing library we expect a higher representation of members with higher
LNA content in the subsequent rounds.
*noget om at Joyce lavede in vitro evolution på et ribozym og fandt katalytisk aktivititet med kun 3
(rogers/joyce 1999) og 2! (reader/joyce 2002) nukleotider.
This reduction in pool size need not be detrimental
One may ask whether the changes that we see in pool composition are due to the use of LNA or whether it is a generic effect that occurs regardless of the type nucleic acids involved. Attempts to run deoxyribonucleotide-only amplification/regenerations in parallel failed after 1-2 rounds (data not shown). We speculate that the conditions that we use for primer extension and/or PCR may be suboptimal for precise and efficient propagation of DNA-only libraries.
18/23
240
245
250
255
Conclusion
We have previously established that Phusion DNA polymerase can use an LNA-containing strand as
Here, we develop on these techniques and show how the actions of both Phusion and KOD (XL) DNA polymerase may be used to generate, amplify and regenerate a DNA library containing either LNA adenosine or LNA thymidine. This is paramount if our goal of being able to create LNA-containing aptamers by in vitro selection is to be achieved. We speculate that the development of such ‘native’ LNA aptamers would offer better structural stability, smaller size, and increased resistance towards
We are currently in the process of performing in vitro selection against several protein and peptide targets. Rather than selecting DNA aptamers and modifying them ‘post SELEX’ we intent to obtain aptamers in which the selection process itself directs the incorporation of LNA moieties. We cordially invite anyone interested in collaborating to contact us.
Acknowledgements
The authors wish to acknowledge Lykke H. Hansen for excellent technical assistance.
H.D. and L.H.L were funded by a stipend from the Danish Agency for Science, Technology and Innovation
(FTP). The Nucleic Acid Center is funded by the Danish National Research Foundation.
19/23
260
265
270
275
280
285
290
Experimental
Oligonucleotides
Phusion PCR with LNA ATP, template and primers: 5′-GGTCTGGTCCACACCCAGCCGCCaCCCaGGGaCG-
CaGCCaGGCaCGGCGGGCCTATAGTGAGTCGTATTA (lowercase is LNA), 5′-GGTCTGGTCCACACCCAG, 5′-
TAATACGACTCACTATAGGCC. LNA incorporation by asymmetric PCR, template and primer: (LNA ATP) 5′-
GGACAGGACCACACCCAGVVVVVVVTVVVVVVTVVVVVVTVVVVVVTVVVVVVGGCCAAAAGAGAGACGAAA,
5′-TTTCGTCTCTCTTTTGGCC; (LNA TTP) 5′-GGACAGGACCACACCCAGABBBBABBBABBBABBBABBBABBB-
ABBBBGGCCTTTTGTGTGTCGTTT, 5′-AAACGACACACAAAAGGCC; *** (LNA GTP) 5′-GGACAGGACCAC-
ACCCAGDDDDDDDCDDDDDDCDDDDDDCDDDDDDCDDDDDDGGAAGGTTGTGTGTAGTTG, 5′-
CAACTACACACAACCTTCC; (LNA m5-CTP) 5′-GGACAGGACCACACCCAGHHHHHHHGHHHHHHGH-
HHHHHGHHHHHHGHHHHHHCACCTTCCATACATCATCC, 5′-GGATGATGTATGGAAGGTG ***. (‘V’ is A/C/G,
‘B’ is C/G/T, ‘D’ is A/G/T, and ‘H’ is A/C/T). N40 library and primers: 5′-GCTCTGTCGTCGTGGTCGGTCC-
(N
40
)-GGCCTTTTGTGTGTCGTTT, 5′-GCTCTGTCGTCGTGGTCGGTCC, 5′-AAACGACACACAAAAGGCC.
Whenever appropriate, chemically synthesized primers with 5 ′ phosphate, 5 ′ 6-carboxyfluorescein
(FAM), or 5 ′ dual biotinylation were used. Radiolabeling was with T4 polynucleotide kinase and 32 P-γ-ATP
[12]. All DNA oligomers were synthesized by Eurofins MWG Operon, except for the dual biotinylated
capture oligomers, which were from Integrated DNA Technologies. LNA oligomers were synthesized inhouse at the Nucleic Acid Center, Department of Physics and Chemistry.
Polymerase chain reaction (PCR)
0.5 nM template (all-DNA or LNA-containing single-stranded DNA) and 0.5 µM of each primer were combined in 1× Phusion HF buffer with 200 µM of each deoxyribonucleotide triphosphate and 0.04 units/µl Phusion DNA polymerase (Finnzymes). PCR conditions were typically: 98 °C/5 min., 20 cycles (98
°C/5 s, 53 °C/10 s, 72 °C/5 min.), 4 °C/hold. Products were resolved by agarose gel electrophoresis.
Lambda exonuclease digestion
Double-stranded DNA was prepared with a 5 ′ -phosphorylated primer and a 5 ′ 6-carboxyfluoresceinlabeled primer. Digestion with lambda exonuclease (New England Biolabs) was with 6.7 units/µg doublestranded DNA (50 ng/µl final DNA concentration) at 37 °C for 25 min. The reaction was stopped by quenching with addition of 1 vol. 50 mM EDTA or by heating to 75 °C for 15 min. Full digestion was verified by agarose gel electrophoresis and ethidium bromide staining. Fluorescence scanning was on a
Typhoon Trio system (GE Healthcare).
Asymmetric PCR
Reaction conditions were: 1×KOD XL buffer, 3 mM MgSO
4
, 0.2 mg/ml BSA, 0.2 u/µl KOD XL DNA polymerase (Novagen), 2.5 µM primer, *TEMPLATE; ENTEN I NANOGRAM ELLER (FOR LASSES ASYPCR-
FIGURER) I nM!*, and 0.25 mM of each nucleotide triphosphate as required. Thermocycling was: 95°/1 min., 15 cycles (95°/15 s, 60°/35 s, 72°/5 min), 4°/hold. Reactions were quenched with 8.3 mM (final)
EDTA and stored at −20 °C.
20/23
295
300
305
310
315
320
325
330
Oligonucleotide capture on magnetic beads
Primer extension products were slow-annealed over 30 min. or more with at least 2-fold molar excess of dual biotinylated oligomer complementary to the products’ 3 ′ end. The solution was adjusted to 1×B&W
(according to manufacturer’s instructions) and combined with Dynabeads M-280 Streptavidin
(Invitrogen) washed twice in 1×B&W buffer. The number of beads corresponded to an excess of biotin binding sites. After 15 min. with gentle agitation the beads were washed first in 1×B&W, then in 1×SSC, before heat-elution of the non-biotinylated species into 40 µl 1×SSC (75 °C, 5 min.).
Digestion assay
Ca. 1.5 pmol of LNA-containing strands isolated after 1, 2 or 3 rounds of amplification and regeneration of the N40 library were 5 ′ -radiolabelled. Full-length oligomers were purified from a 6% denaturing polyacrylamide gel and eluted into 100 µl water. Sample volumes were then adjusted to achieve equivalent specific activity. Similarly, the crude DNA library and an LNA A oligomer (5 ′ -GGTCTGGTCC-
ACACCCAGCCGCCaCCCaGGGaCGCaGCCaGGCaCGGCGGGCCTATAGTGAGTCGTATTA; lower case is LNA A) were 5 ′ -radiolabelled to 7.5 nM. 5 µl of each oligomer was then incubated at 72 °C in digestion buffer
(1× KOD buffer #2, 3 mM MgSO
4
, 0.2 mg/ml BSA) with or without 0.2 units KOD DNA polymerase
(Novagen) (10 µl final volume). 1.5 µl samples were drawn at intervals, quenched in 1 vol. ice-cold 95% formamide/50 mM EDTA and resolved on 13% denaturing polyacrylamide gels. Autoradiography was with the PhosphoImager system (GE Healthcare), and line densitometry was with spline curves (width:
30) in ImageJ 1.44j (NIH, http://rsb.info.nih.gov/ij/). Data pairs (i.e. with/without polymerase) were matched to achieve identical overall intensity and all pairs were normalized according to peak values
(Excel 2010, Microsoft).
LC/MS analysis
Phusion DNA polymerase was used to generate double-stranded DNA from LNA-containing templates.
One primer was dual biotinylated and was used for oligonucleotide capture. The eluted single-stranded
DNA libraries were digested with nuclease P1 (Roche) , phosphodiesterase I (Sigma), and alkaline
phosphatase (Sigma) [10], and the non-nucleoside content was methanol-precipitated. The supernatant
was extracted, dried and re-dissolved in 0.1% formic acid/1 µM 5-methylcytidine (m5C). Nucleosides were quantified by nano-chip LC/MS on an Agilent 6340 Ion Trap using a modified method according to
[13]. *STUFF ‘BOUT THE GRADIENT GOES HERE* Samples were injected at both 1:9 and 1:99 dilutions for
improved dynamic range. We used m5C as internal standard for sample normalization. Nucleoside ratios were subsequently normalized to the theoretical composition of the initial library in order to account for differences in ionization efficiency of the various nucleosides. Calculations were done in Excel 2010
(Microsoft).
Sequencing
Phusion DNA polymerase was used to generate double-stranded DNA from LNA-containing templates.
The DNA was then 5 ′ -phosphorylated and blunt-end cloned into SmaI-digested pUC18 or pUC19; this offers multiple inserts per plasmid. The plasmids were transformed into E. coli TOP10. Plasmids from restreaked colonies were purified (Miniprep, Qiagen), and cycle sequencing at Eurofins MWG Operon
(Germany) was with the ‘M13 rev (−49)’ primer. All extracted sequences are available in Supplementary
21/23
22/23
335
340
345
350
355
360
365
370
References
1.
2.
3.
4.
5.
6.
Veedu, R.N., B. Vester, and J. Wengel, In vitro incorporation of LNA nucleotides.
Nucleosides
Nucleotides Nucleic Acids, 2007. 26(8-9): p. 1207-10.
Veedu, R.N., B. Vester, and J. Wengel, Novel applications of locked nucleic acids.
Nucleic Acids
Symp Ser (Oxf), 2007(51): p. 29-30.
Veedu, R.N., B. Vester, and J. Wengel, Enzymatic incorporation of LNA nucleotides into DNA strands.
Chembiochem, 2007. 8(5): p. 490-2.
Veedu, R.N., B. Vester, and J. Wengel, Polymerase chain reaction and transcription using locked nucleic acid nucleotide triphosphates.
J Am Chem Soc, 2008. 130(26): p. 8124-5.
Veedu, R.N., B. Vester, and J. Wengel, Efficient enzymatic synthesis of LNA-modified DNA duplexes using KOD DNA polymerase.
Org Biomol Chem, 2009. 7(7): p. 1404-9.
Kuwahara, M., et al., Systematic analysis of enzymatic DNA polymerization using oligo-DNA templates and triphosphate analogs involving 2',4'-bridged nucleosides.
Nucleic Acids Res, 2008.
36(13): p. 4257-65.
7.
8.
Doessing, H., et al., Synthesis and isolation of a single-stranded DNA library with multiple LNA-A modifications .
Nishioka, M., et al., Long and accurate PCR with a mixture of KOD DNA polymerase and its exonuclease deficient mutant enzyme.
J Biotechnol, 2001. 88(2): p. 141-9.
9. Little, J.W., An exonuclease induced by bacteriophage lambda. II. Nature of the enzymatic reaction.
J Biol Chem, 1967. 242(4): p. 679-86.
10. Crain, P.F., Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry.
Methods Enzymol, 1990. 193: p. 782-90.
11. Veedu, R.N. and J. Wengel, Locked nucleic acid nucleoside triphosphates and polymerases: on the way towards evolution of LNA aptamers.
Mol Biosyst, 2009. 5(8): p. 787-92.
12. Maniatis, T., E.F. Fritsch, and J. Sambrook, Molecular cloning : a laboratory manual . Manual for genetic engineering. Vol. 1. 1982, Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory.
13. Giessing, A.M., et al., Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria.
RNA, 2009.
15(2): p. 327-36.
14. Thompson, J.D., D.G. Higgins, and T.J. Gibson, CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice.
Nucleic Acids Res, 1994. 22(22): p. 4673-80.
15. Waterhouse, A.M., et al., Jalview Version 2--a multiple sequence alignment editor and analysis workbench.
Bioinformatics, 2009. 25(9): p. 1189-91.
16. Crooks, G.E., et al., WebLogo: a sequence logo generator.
Genome Res, 2004. 14(6): p. 1188-90.
23/23
Supplementary
Figure S1: Sequencing results from PCR amplification of an LNA-containing library with Phusion High
Fidelity DNA polymerase.
LNA library sequence: 5 ′ -GGACAGGACCACACCCAGaBBBBaBBBaBBBaBBBaBBBaBBBaBBBBGGCCTTTT-
GTGTGTCGTTT-3 ′ , where lowercase ‘a’ is LNA A and ‘B’ is G, C, or T. Below, the flanking primer sequences (determined by local sequence alignment) are highlighted in capital letters. Some clones yielded the reverse complementary sequence due to the blunt-end cloning procedure used.
1.
GGACAGGACCACACCCAGagtgaccgaccgaccgacggatttagcgcGGCCTTTTGTGTGTCGTTT
2.
AAACGACACACAAAAGGCCcgcatccctaactccatacctcgatccgtCTGGGTGTGGTCCTGTCC
3.
GGACAGGACCACACCCAGattgaggcactgatgtaggtatggacctgGGCCTTTTGTGTGTCGTTT
4.
GGACAGGACCACACCCAGacttagggatgcacccacttatttatttgGGCCTTTTGTGTGTCGTTT
5.
GGACAGGACCACACCCAGatggacgcatttacttactgatctatctcGGCCTTTTGTGTGTCGTTT
6.
AAACGACACACAAAAGGCCcccatacgtgaatcaataactgggtgcgtCTGGGTGTGGTCCTGTCC
7.
GGACAGGACCACACCCAGagggaccgatggactgattgatgtagcggGGCCTTTTGTGTGTCGTTT
8.
GGACAGGACCACACCCAGacgcattgactcacgcactcacccacttggGGCCTTTTGTGTGTCGTTT
9.
GGACAGGACCACACCCAGacccatgcatctactgatcgatttagtgcGGCCTTTTGTGTGTCGTTT
10.
AAACGACACACAAAAGGCCcggatagatacctcaataactgcctacatCTGGGTGTGGTCCTGTCC
11.
AAACGACACACAAAAGGCCacgctcaatccatcgctcgctacctgcctCTGGGTGTGGTCCTGTCC
12.
GGACAGGACCACACCCAGagggattcattgattgacccaggtagtggGGCCTTTTGTGTGTCGTTT
13.
AAACGACACACAAAAGGCCcaggtgagtaggtccctccctccatcggtCTGGGTGTGGTCCTGTCC
14.
AAACGACACACAAAAGGCCccactcgctggatcgctagctaagtccctCTGGGTGTGGTCCTGTCC
15.
GGACAGGACCACACCCAGagggagtgatgtagggaggcagggagtgtGGCCTTTTGTGTGTCGTTT
16.
GGACAGGACCACACCCAGgagttacgcacgcatctaggcatccaccttGGCCTTTTGTGTGTCGTTT
17.
GGACAGGACCACACCCAGagctactgattcatgtatgggatccatgtgGGCCTTTTGTGTGTCGTTT
18.
GGACAGGACCACACCCAGatggagccattgatctagctacggactttGGCCTTTTGTGTGTCGTTT
19.
AAACGACACACAAAAGGCCcacatacatacctccctcactccctccgtCTGGGTGTGGTCCTGTCC
20.
AAACGACACACAAAAGGCCcaggtcgctaactggatccataagtggatCTGGGTGTGGTCCTGTCC
21.
GGACAGGACCACACCCAGacctattcacccagcgagccacctactgtGGCCTTTTGTGTGTCGTTT
22.
AAACGACACACAAAAGGCCgaaatgactcaataagtgcctgcgtaaatCTGGGTGTGGTCCTGTC
23.
GGACAGGACCACACCCAGagtgatttattcacgtagtcattgagctcGGCCTTTTGTGTGTCGTTT
24.
GGACAGGACCACACCCAGattcagcgagcgagggactcatttagtccGGCCTTTTGTGTGTCGTTT
25.
GGACAGGACCACACCCAGagctacccacgcaggtatgtatcaggctGGCCTTTTGTGTGTCGTTT
26.
AAACGACACACAAAAGGCCaacctcactgaatgcatagctccctgcatCTGGGTGTGGTCCTGTC
27.
AAACGACACACAAAAGGCCcggatcgctacctagctagctccctccgtCTGGGTGTGGTCCTGTCC
28.
GGACAGGACCACACCCAGacgtaccgatgcatttatctacccagtgtGGCCTTTTGTGTGTCGTTT
29.
GGACAGGACCACACCCAGactcagttaccgacgtaccgatttagctgGGCCTTTTGTGTGTCGTTT
30.
GGACAGGACCACACCCAGagtcagggagccacctaggcacttagttgGGCCTTTTGTGTGTCGTTT
31.
AAACGACACACAAAAGGCCcacatagatccatcgatgactccctaaatCTGGGTGTGGTCCTGTCC
32.
GGACAGGACCACACCCAGatggatctaggtagctatttaggcaggctGGCCTTTTGTGTGTCGTTT
S1








