Noter til 2 timer eksamen - sociologisk
advertisement

Noter til 2 timer eksamen
Appendix
Grundlæggende formler:
Population
Stikprøve
k
Middelværdi
𝑛
μ/E(X) = ∑ xj f(xj )
𝑌̅ = 𝑛
j=1
−1
∑ 𝑌𝑖
𝑖=1
𝑛
Varians
2
𝑉𝑎𝑟(𝑋)/𝜎 = 𝐸(𝑋
2)
−𝜇
1
𝑆 =
∑(𝑌𝑖 − 𝑌̅)2
𝑛−1
2
2
𝑖=1
Standard afvigelse/fejl
Kovarians
Korrelationskoefficient
95 % kofidensinterval for
stor n (<120)
𝑠𝑑(𝑋)/𝜎 = √𝑉𝑎𝑟(𝑋)
𝐶𝑜𝑣(𝑋, 𝑌)
= 𝐸[(𝑋 − 𝜇𝑋 )(𝑌 − 𝜇𝑌 )]
𝜎𝑋𝑌
𝐶𝑜𝑣(𝑋, 𝑌)
𝐶𝑜𝑟𝑟(𝑋, 𝑌) =
, [−1; 1]
𝑠𝑑(𝑋) ∙ 𝑠𝑑(𝑌)
𝑠𝑒 = √𝑆 2
𝑛
𝑆𝑋𝑌
1
=
∑(𝑋𝑖 − 𝑋̅)(𝑌𝑖 − 𝑌̅)
𝑛−1
𝑖=1
𝑅𝑋𝑌 =
𝑆𝑋𝑌
𝑆𝑋 𝑆𝑌
𝑦̅ ± 1,96 ∙ 𝑠𝑒(𝑦̅)
Store bogstaver: estimatorer
Små bogstaver: estimater
∑(𝑥𝑖 − 𝑥̅ ) = 0
Dvs. hvis hver observation trækkes fra gennemsnittet, så er summen af disse forskelle lig nul.
Finite sample: the properties hold for a sample of any size, no matter how small or large
Unbiasedness: An estimator, W of Ѳ, is an unbiased estimator if: E(W) = Ѳ, for all possible values
of Ѳ.
Efficiency: If W1 and W2 are two unbiased estimators of Ѳ, W1 is efficient relative to W2 when
Var(W1) ≤ Var(W2) for all Ѳ
Consistency: Let Wn be an estimator of Ѳ based on a sample Y1, Y2, … Yn og size n. Then Wn is a
consistent estimator of Ѳ if for every ε > 0, P(|Wn – Ѳ| > ε) → 0 as n → ∞
1
Chapter 2 – The Simple Regression Model
SLR1 – zero conditional mean assumption
E(u|x) = E(u) = 0
SLR1 gives us the population regression function (PRF): E(y|x) = β0+ β1x
The sample regression function: 𝑦̂ = 𝛽̂0 + 𝛽̂1 𝑥
Ordinary least squares estimates (OLS):
Fitted value: 𝑦̂𝑖 = 𝛽̂0 + 𝛽̂1 𝑥𝑖
Residuals: 𝑢̂𝑖 = 𝑦𝑖 − 𝑦̂𝑖
𝛽̂0 = 𝑦̅ − 𝛽̂1 𝑥̅
𝑛
∑ (𝑥 −𝑥̅ )(𝑦𝑖 −𝑦̅)
𝛽̂1 = 𝑖=1𝑛 𝑖
2
∑𝑖=1(𝑥𝑖 −𝑥̅ )
(𝑢̂𝑖 is not the same as ui – while the residuals are
computed from the data, the errors are never observable)
The 3 most important algebraic properties of OLS residuals:
1. The sum, and therefore the sample average of the OLS residuals, is zero ∑𝑛𝑖=1 𝑢̂𝑖 = 0
2. The sample covariance between the independent variables and the OLS residuals is zero
∑𝑛𝑖=1 𝑥𝑖 𝑢̂𝑖 = 0
3. The point (𝑥̅ , 𝑦̅) is always on the OLS regression line
SST, SSE, SSR and R2:
Total sum of squares (SST):
Explained sum of squares (SSE):
Residual sum of squares (SSR):
∑𝑛𝑖=1(𝑦𝑖 − 𝑦̅)2
∑𝑛𝑖=1(𝑦̂𝑖 − 𝑦̅)2
∑𝑛𝑖=1 𝑢̂𝑖2
total sample variation in yi
sample variation in 𝑦̂𝑖
sample variation in 𝑢̂𝑖
SST = SSE + SSR
R2 = SSE/SST = 1-SSR/SST
Summary of functional forms involving logarithms:
Model
Level-level
Level-log
Log-level
Log-log
Dependent variable
y
y
log(y)
log(y)
Independent variable
x
log(x)
x
log(x)
Interpretation of β1
Δy = β1 Δx
Δy = (β1/100)%Δx
%Δy = (100β1)Δx
%Δy = β1%Δx
Homoskedasticity:
Because Var(u|x) = E(u2|x)-[E(u|x)]2 and E(u|x) = 0, σ2 = E(u2|x), which means σ2 is the
unconditional expectation of u2. Therefore, σ2 = E(u2) = Var(u), because E(u) = 0
Var(u|x) = Var(y|x) - heteroskedasticity is present whenever Var(y|x) is a function of x.
σ2 = the error variance
σ = the standard deviation of the error
2
Variance and standard deviation of the estimates under SLR1-5:
𝑉𝑎𝑟(𝛽̂1 ) =
𝜎2
2
∑𝑛
𝑖=1(𝑥𝑖 −𝑥̅ )
𝜎2
=
𝑉𝑎𝑟(𝛽̂0 ) =
𝑆𝑆𝑇𝑥
σ2 ↑ --- Var(β1) ↑
variability in xi ↑ --- Var(β1) ↓
→
2
𝜎 2 𝑛−1 ∑𝑛
𝑖=1 𝑥𝑖
2
∑𝑛
𝑖=1(𝑥𝑖 −𝑥̅ )
n ↑ --- Var(β1) ↓
Estimation of the standard error of 𝛽̂1:
̂
̂
𝜎
𝜎
𝑠𝑒(𝛽̂1 ) =
=
1/2
√𝑆𝑆𝑇𝑥
2
(∑𝑛
𝑖=1(𝑥𝑖 −𝑥̅ ) )
Error variance and standard error of the regression:
Estimation of the error variance, 𝜎̂ 2 / 𝑠 2 :
𝜎̂ 2 =
1
(𝑛−2)
∑𝑛𝑖=1 𝑢̂𝑖2 =
𝑆𝑆𝑅
(𝑛−2)
Estimation of the standard error of the regression: 𝜎̂ = √𝜎̂ 2
Chapter 3 – Multiple Regression Analyses: Estimation
The Linear Regression Model in Matrix Form:
yt =
xt
β
+ ut
, t = 1,2, …, n.
(nx1) (nx(k+1)) ((k+1)xn) (nx1)
[row x column]
̂ = (X’X)-1 (X’y)
𝜷
The OLS model
OLS fitted/predicted values: 𝑦̂ = 𝛽̂0 + 𝛽̂1 𝑥1 + 𝛽̂2 𝑥2 + ⋯ + 𝛽̂𝑘 𝑥𝑘
OLS written in terms of change: ∆𝑦̂ = 𝛽̂1 ∆𝑥1 + 𝛽̂2 ∆𝑥2 + ⋯ + 𝛽̂𝑘 ∆𝑥𝑘
(Notice the intercept is out)
OLS written in terms of change of the coefficient x1 holding all other independent variables fixed:
∆𝑦̂ = 𝛽̂1 ∆𝑥1
OLS written in terms of change of the coefficients x1 and x2 (same unites) holding all other
independent variables fixed: ∆𝑦̂ = 𝛽̂1 ∆𝑥1 + 𝛽̂2 ∆𝑥2
3
First order conditions for the OLS estimators
The OLS residuals are defined as in the simple regression case: (𝑢̂𝑖 = 𝑦𝑖 − 𝑦̂𝑖 ) and have the same
properties:
1. The sum, and therefore the sample average of the OLS residuals, is zero 𝑦̅ = 𝑦̅̂
2. The sample covariance between the independent variables and the OLS residuals is zero
∑𝑛𝑖=1 𝑥𝑖𝑗 𝑢̂𝑖 = 0
3. The point (𝑥̅1 , 𝑥̅2 , … , 𝑥̅𝑘 , 𝑦̅) is always on the OLS regression line
Assumptions: MLR1-5
MLR1: Linear in parameters – The model in the population can be written as:
𝑦 = 𝛽0 + 𝛽1 𝑥1 + 𝛽2 𝑥2 + ⋯ + 𝛽𝑘 𝑥𝑘 + 𝑢
MLR2: Random sample – we have a random sample of n observations, {(xi1, xi2, …, xik, y): i=1,2…,
n}, following the population model in MLR1
(random sample = i.i.d – independent identical distributed)
MLR3: No perfect collinearity – in the sample, none of the independent variables is constant, and
there are no exact linear relationships among the independent variables (if MLR3 is not met the
model suffers from perfect collinearity)
MLR4: Zero conditional mean – the error u has an expected value of zero given any values of the
independent variables. E(u|x1, x2, …, xk) = 0
3 ways for MLR4 to be violated: 1) the functional relationship between the independent variables
and the dependent is misspecified, 2) omitting an important variable, 3) measurement error in an
explanatory variable
Under MLR1-4 the OLS estimators are unbiased estimators of the population parameters: 𝐸(𝛽̂𝑗 ) =
𝛽𝑗 , 𝑗 = 0,1, … , 𝑘,
(An estimate cannot be unbiased, but the procedure by which the estimate is obtained can be
unbiased when we view the procedure as being applied across all possible random samples.)
MLR5: homoskedasticity – the error u has the same variance given any of the explanatory
variables. Var(u|x1, x2, …, xk) = σ2
MLR1-5 = The Gauss-Markov assumptions
MLR1 and MLR4 gives: 𝐸(𝑦|𝒙) = 𝛽0 + 𝛽1 𝑥1 + 𝛽2 𝑥2 + ⋯ + 𝛽𝑘 𝑥𝑘
4
Estimation of the variances and standard errors in multiple regression analysis:
Estimation of the sampling variances of the slope estimators
𝑣𝑎𝑟(𝛽̂𝑗 ) =
̂2
𝜎
𝑆𝑆𝑇𝑗 (1−𝑅𝑗2 )
Where 𝑆𝑆𝑇𝑗 = ∑𝑛𝑖=1(𝑥𝑖𝑗 − 𝑥̅𝑗 )2 is the total sample variation in xj and 𝑅𝑗2 is the R-squared from
regressing xj on all other independent variables. (Valid under MLR1-5)
The sample error variance 𝜎̂ 2 ↑ --- 𝑣𝑎𝑟(𝛽̂𝑗 ) ↑
(Reduce σ2 by adding more explanatory variables)
The total sample variation in xj SSTj ↑ --- 𝑣𝑎𝑟(𝛽̂𝑗 ) ↓
(Increase sample variation in xj by increasing the sample size)
The linear relationship between among the independent variables 𝑅𝑗2 ↑ --- 𝑣𝑎𝑟(𝛽̂𝑗 ) ↑
(Avoid too much multicollinearity – e.g. collect more data)
Omitted variables only cause bias if they are correlated with independent variables in the modeltherefore including irrelevant variables is not a good idea because it will most likely make the
multicollinairty problem bigger (𝑣𝑎𝑟(𝛽̂𝑗 ) ↑)
If omitted variables are correlated with independent variables in the model they should off cause
be included to avoid bias.
The estimate of the standard variance of the regression: 𝜎̂ 2 = 𝑆𝑆𝑅/(𝑛 − 𝑘 − 1)
The estimate of the standard error of the regression: 𝜎̂ = √𝜎̂ 2
The estimate of the standard error of 𝛽̂𝑗 under MLR5: 𝑠𝑒(𝛽̂𝑗 ) = 𝜎̂/[𝑆𝑆𝑇𝑗 (1 − 𝑅𝑗2 )]1/2
Under MLR1-5 OLS gives unbiased estimation of σ2, 𝐸(𝜎̂ 2 ) = 𝜎 2
Normality assumption – MLR6
MLR6: Normality - The population error u is independent of the explanatory variables x1, x2, …, xk,
and is normally distributed with zero mean and variance σ2: ~ u Normal(0, σ2).
(strong assumption that is problematic in several cases – e.g. if y takes on only a few values)
MLR1-6: The classical linear model assumptions (CLM)
Under CLM1-6 𝛽̂𝑗 ~𝑁𝑜𝑟𝑚𝑎𝑙[𝛽𝑗 , 𝑉𝑎𝑟(𝛽̂𝑗 )] therefore (𝛽̂𝑗 − 𝛽𝑗 )/𝑠𝑑(𝛽̂𝑗 )~𝑁𝑜𝑟𝑚𝑎𝑙(0,1)
In addition any linear combination of the 𝛽̂0 , 𝛽̂1 , 𝛽̂2 , … , 𝛽̂𝑘 is also normally distributed.
5
Assumptions
MLR1-4: OLS is LUE
MLR1-5: OLS is BLUE
CLM1-6: OLS estimators has the smallest variance among all unbiased estimators – not only in
comparison to linear estimators (MLR6 is a strong assumption that’s only necessary with small
samples to know the sampling distribution for inference)
Bias
Omitted variable bias – the simple case
- Bias when omitting the explanatory variable x2 from the model y = β0+ β1x1 + β2x2 + u
𝐵𝑖𝑎𝑠(𝛽̃1 ) = 𝐸(𝛽̃1 ) − 𝛽1 = 𝛽2 𝛿̃1
𝛽̃1 comes from the underspecified model without x2
𝛽1 and 𝛽2 comes from the specified model with x2
𝛿̃1 is the slope from the simple regression of x2 on x1 (if x2 on x1 are uncorrelated in the sample
then 𝛽̃1 is unbiased)
Summary of bias in 𝛽̃1when x2 is omitted in estimating equation y = β0+ β1x1 + β2x2 + u
Corr(x1, x2) > 0
Corr(x1, x2) < 0
β2 > 0
Positive bias
Negative bias
β2 < 0
Negative bias
Positive bias
̃
̃
Upward bias in 𝛽1:
𝐸(𝛽1 ) > 𝛽1
Downward bias in 𝛽̃1:
𝐸(𝛽̃1 ) < 𝛽1
Biased toward zero: the case where 𝐸(𝛽̃1 ) is closer to zero than β1
Omitted variable bias – the more general case with multiple regressors in the estimated model
- Bias when omitting the explanatory variable x3 from the model y = β0+ β1x1 + β2x2 + β3x3 u
Assume that x2 is uncorrelated with x1 and x3 – then we can study the bias in x1 as if x2 were absent
from the model. This means we can use the above equation and table (now just with x 3 instead of
x2)
6
Chapter 4 – Multiple Regression Analysis: Inference
t and F tests and confidence intervals
Under CLM1-6 or with large samples:
𝑡𝛽̂𝑗 = (𝛽̂𝑗 − 𝛽𝑗 )/𝑠𝑒(𝛽̂𝑗 )~𝑡𝑛−𝑘−1
where k +1 is the number of unknown parameters in the population model (k slope parameters
and the intercept)
𝐹=
(𝑆𝑆𝑅𝑟 −𝑆𝑆𝑅𝑢𝑟 )/𝑞
𝑆𝑆𝑅𝑢𝑟 /(𝑛−𝑘−1)
~𝐹(𝑞, 𝑛 − 𝑘 − 1)
The restricted model (r) always has fewer parameters than the unrestricted model (ur).
q is the number of exclusion restrictions to test (=df r - dfur)
𝐹=
(𝑅 2 𝑢𝑟 −𝑅 2𝑟 )/𝑞
1−𝑅 2 𝑢𝑟 /(𝑛−𝑘−1)
~𝐹(𝑞, 𝑛 − 𝑘 − 1)
Since SSRr can be no smaller than SSRur, the F statistic is always nonnegative
The F statistic is often useful for testing exclusion of a group of variables when the variables in the
group are highly correlated (when the multicollinearity makes it difficult to uncover the partial
effect)
It can be shown that F statistic for testing exclusion if a single variable is equal to the square of the
corresponding t statistic.
A 95% confidence interval: 𝛽̂𝑗 ∓ 𝑐 ∙ 𝑠𝑒(𝛽̂𝑗 ) where the constant c is the 97,5th percentile in a tn-k.1
distribution.
Testing hypotheses about a single linear combination of the parameters:
H0: β1 = β2 => β1 - β2 = 0
𝑡 = (𝛽̂1 − 𝛽̂2 )/𝑠𝑒(𝛽̂1 − 𝛽̂2 )
2
2
𝑠𝑒(𝛽̂1 − 𝛽̂2 ) = {[𝑠𝑒(𝛽̂1 )] + [𝑠𝑒(𝛽̂2 )] − 2𝐶𝑜𝑣(𝛽̂1, 𝛽̂2 )}1/2
To get the right standard error for the test it is easiest to estimate a new model where we define a
new parameter as the difference between β1 and β2 – do this by including x1 + x2 in the equation
instead of x2 and then the estimate and standard error of x1 can be used for the test - see page
142…
Significance level – the probability of rejecting H0 when it is in fact true (we will mistakenly reject
H0 when it is true 5% of the time)
One sided test: the critical value is the 95th percentile in a t distribution with n-k-1 degrees of
freedom
Two sided test: the critical value is the 97,5th percentile in a t distribution with n-k-1 degrees of
freedom
7
The p-value: the smallest significance level at which the null hypothesis would be rejected. The pvalue is the probability of observing a t statistic as extreme as we did if the null hypothesis is true –
small p-values are evidence against the null.
Chapter 5 – Multiple Regression Analysis: OLS Asymptotics
Consistency = asymptotic unbiased
If an estimator is consistent then the distribution of 𝛽̂𝑗 becomes more and more tightly distributed
around βj as the sample size grows. As n tends to infinity, the distribution of 𝛽̂𝑗 collapses to the
single point βj (e.i. plim 𝛽̂𝑗 = βj)
MLR4’
Zero mean and zero correlation. E(u) = 0 and Cov(xj,u) = 0, for j = 1, 2, …, k.
MLR4’ is weaker than MLR4: MLR4 requires that any function of xj is uncorrelated with u,
MLR4´requires only that each xj is uncorrelated with u.
OLS is biased but consistent under MLR4’ if E(u|x1, …, xk) depends on any of xj
But if we only assume MLR4’ MLR1 need not to represent the population regression model (PRF),
and we face the possibility that some nonlinear function of the xj such as xj2 could be correlated
with the error u. This means that we have neglected nonlinearities in the model that could help us
better explain y; if we knew that we would usually include such nonlinear functions. That is, most
of the time we hope to get a good estimate of the PRF, and so MLR4 (the ‘normal’ one) is natural
(we use MLR’ with IV where we have no interest in modelling PRF).
Inconsistency in the estimators – asymptotic bias
Correlation between u and any of xj causes all of the OLS estimators to be biased and inconsistent
(if the independent variables in the model are correlated – which is usually the case).
Any bias persists as the sample size grows – the problem actually gets worse with more
observations.
The inconsistency in 𝛽̂1(sometimes called the asymptotic bias) is
𝑝𝑙𝑖𝑚𝛽̂1 − 𝛽1 = 𝑐𝑜𝑣(𝑥1 , 𝑢)/𝑣𝑎𝑟(𝑥1 )
Because var(x1) is positive the inconsistency in 𝛽̂1is positive if 𝑐𝑜𝑣(𝑥1 , 𝑢) is positive and negative if
𝑐𝑜𝑣(𝑥1 , 𝑢) is negative.
Asymptotic analog of omitted variable bias – the simple case:
Suppose the true model is y = β0+ β1x1 + β2x2 + u and we omit x2. Then
𝑝𝑙𝑖𝑚(𝛽̃1) = 𝛽1 + 𝛽2 𝛿1
𝛿1 = 𝑐𝑜𝑣(𝑥1 , 𝑥2 )/𝑣𝑎𝑟(𝑥1 )
𝛽̃1 comes from the underspecified model without x2
𝛽1 and 𝛽2 comes from the specified model with x2
Asymptotic normality
8
Even though yi are not from a normal distribution (MLR6), under MLR1-5 we can use the central
limit theorem to conclude that OLS estimators satisfy asymptotic normality, which means they are
approximately normally distributed in large enough sample sizes.
𝜎̂ 2 is a consistent estimator of σ2 – an asymptotic analysis can know show that 𝑣𝑎𝑟(𝛽̂𝑗 ) shrinks to
zero at the rate of 1/n; this is why a large sample size are better. The standard error can be
expected to shrink at a rate that is the inverse of the square root of the sample size (𝑐𝑗 /√𝑛 where
cj is a positive constant that does not depend on the sample size.
Lagrange multiplier (LM) statistic for q exclusion restrictions:
(Works under MLR1-5 with large sample. Same hypothesis as with F tests)
o
o
o
o
Regress y on the restricted set of independent variables and save the residuals, 𝑢̃
Regress 𝑢̃ on all of the independent variables and obtain the R-squared, 𝑅𝑢2
Compute LM = 𝑛 ∙ 𝑅𝑢2
Compare LM to the appropriate critical value in a 𝜒𝑞2 distribution
Auxiliary regression
- a regression that is used to compute a test statistic but whose coefficients are not of direct
interest.
Chapter 6 – Multiple Regression Analysis: Further Issues
Changing the units of measurement:
If xj is multiplied by c, its coefficient is divided by c. If the dependent variable is multiplied by c, all
OLS coefficients are multiplied by c.
Using Logarithmic Functional Forms:
Log-level model: As the change in log(y) becomes larger and larger, the approximation
%Δy~100·Δlog(y) becomes more and more inaccurate. The exact percentage change in the
predicted y is given by: %Δŷ = 100 ∙ [exp(β̂ 2 ∆x2 ) − 1]
Simple using the coefficient (multiplied by 100) gives us an estimate that is always between the
absolute value of the estimates for an increase and a decrease. If we are especially interested in
an increase or a decrease we can use the calculation based on the equation above.
Reasons for using log models:
When y>0, models using log(y) as the dependent variable often satisfy CLM assumptions more
closely than models using the level of y. Moreover taking log usually narrows the range of the
variable, which makes estimates less sensitive to outlying or extreme observations on the
dependent or independent variables.
When a variable is a positive dollar amount or a large positive whole value the log is often taken.
Variables that are measured in years usually appear in their original form
9
A variable that is a proportion or percent usually appear in level from, but can also appear in log
form (here we will have a percentage point change)
Log cannot be used if a variable takes on zero or negative values.
Models with quadratics:
If we write the estimated model as: 𝑦̂ = 𝛽̂0 + 𝛽̂1 𝑥 + 𝛽̂2 𝑥 2 + 𝑢 then we have the approximation:
∆𝑦̂ = (𝛽̂1 + 2𝛽̂2 𝑥)∆𝑥 , for ‘small’ Δx
This say that the slope of the relationship between x and y depend on the value of x. 𝛽̂1 can be
interpreted as the approximate slope in going from x=0 to x=1.
Turning point
𝑥 ∗ = |𝛽̂1 /2𝛽̂2 |
𝛽̂1 is positive and 𝛽̂2 is negative – x has a diminishing effect on y, parabolic shape
𝛽̂1 is negative and 𝛽̂2 is positive – x has an increasing effect on y, u-shape
𝛽̂1 and 𝛽̂2 have the same sign – there is no turning point for values x > 0. Both positive - the
smallest expected value of y is at x = 0 and increases in x always have a positive effect on y. Both
negative – the largest expected value of y is at x = 0 and increases in x always have a negative
effect on y.
Models with interaction terms:
If we write the estimated model as: 𝑦̂ = 𝛽̂0 + 𝛽̂1 𝑥1 + 𝛽̂2 𝑥2 + 𝛽̂3 𝑥1 𝑥2 + 𝑢 then 𝛽̂2 is the partial
effect of x2 on y when x1 = 0
∆𝑦̂
= 𝛽̂2 + 𝛽̂3 𝑥1
∆𝑥2
To estimate the effect of x2 plug in interesting values of x1 – e.g. the mean.
Predicting y when log(y) is the dependent variable:
𝜎̂ 2
̂
𝑦̂ = exp ( ) exp(𝑙𝑜𝑔𝑦)
2
Chapter 7 – Multiple Regression Analysis with Qualitative Information: Binary/dummy Variables
Difference in intercept – dummy variable:
𝑤𝑎𝑔𝑒
̂ = 𝛽̂0 + 𝛽̂1 𝑒𝑑𝑢𝑐 + 𝛽̂2 𝑓𝑒𝑚𝑎𝑙𝑒 + 𝑢 Then the intercept for males is 𝛽̂0 and the intercept for
females is 𝛽̂0 + 𝛽̂2
If the regression model needs to have different intercept for, say, g groups or categories, we need
to include g-1 dummy variables in the model along with an intercept. The intercept for the base
group is the overall intercept in the model, and the dummy variable for a particular group
represent the estimated difference in intercepts between that group and the base group.
10
Interactions among dummy variables:
𝑤𝑎𝑔𝑒
̂ = 𝛽̂0 + 𝛽̂1 𝑚𝑎𝑟𝑟𝑖𝑒𝑑 + 𝛽̂2 𝑓𝑒𝑚𝑎𝑙𝑒 + 𝛽̂3 𝑓𝑒𝑚𝑎𝑙𝑒 ∙ 𝑚𝑎𝑟𝑟𝑖𝑒𝑑 + 𝑢 Then we can obtain the
estimated wage differential among all four groups, but here we must be careful to plug in the
correct combination of zero and ones. Setting female = 0 and married = 0 correspond to the group
single men, which is the base group since this eliminates all three parameters. We can find the
intercept for married men by setting female = 0 and married = 1.
Difference in slope – interactions between dummy and quantitative variables:
𝑤𝑎𝑔𝑒
̂ = 𝛽̂0 + 𝛽̂1 𝑒𝑑𝑢𝑐 + 𝛽̂2 𝑓𝑒𝑚𝑎𝑙𝑒 + 𝛽̂3 𝑓𝑒𝑚𝑎𝑙𝑒 ∙ 𝑒𝑑𝑢𝑐 + 𝑢 Then 𝛽̂2 measures the difference in
intercepts between women and men, and 𝛽̂3 measures the difference in the return to education
between women and men.
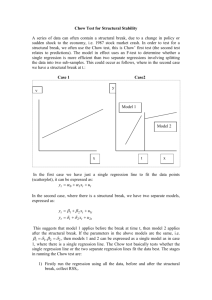
CHOW test - Testing for difference in regression function across groups:
We test the null hypothesis that two populations or groups follow the same regression function,
against the alternative that one or more of the slopes differ across groups.
(can also be done by adding all of the interactions and computing the F statistic)
Chow statistic (e.g. with two groups)
𝐹=
[𝑆𝑆𝑅𝑝𝑜𝑜𝑙𝑒𝑑 − (𝑆𝑆𝑅𝑔𝑟𝑜𝑢𝑝 1 + 𝑆𝑆𝑅𝑔𝑟𝑢𝑜𝑝 2 )] [𝑛 − 𝑛𝑢𝑚𝑏𝑒𝑟 𝑜𝑓 𝑔𝑟𝑜𝑢𝑝𝑠(𝑘 + 1)]
∙
(𝑆𝑆𝑅𝑔𝑟𝑜𝑢𝑝 1 + 𝑆𝑆𝑅𝑔𝑟𝑢𝑜𝑝 2 )
𝑞(𝑘 + 1)
Where n is the total number of observations, k is the number of explanatory variables, q is the
number of dummy variables – 1 (the base group) (with two groups q is zero) and SSRpooled is SSR1 +
SSR2.
A binary dependent variable – the linear probability model:
P(y=1|x)=E(y|x) : the probability of success, that is the probability that y=1, is the same as the
expected value of y.
In the LPM model βj measures the change in the probability of success when xj changes/increases
with one unit, holding other factors fixed.
Var(y|x)=p(x)[1-p(x)]
Chapter 8 – Heteroskedasticity
Heteroskedasticity-robust standard error for βj:
𝑣𝑎𝑟(𝛽̂𝑗 ) =
2 2
∑𝑛
̂𝑖
𝑖=1 𝑟̂𝑖𝑗 𝑢
𝑆𝑆𝑅𝑗2
- useful only with large samples.
The robust std. errors can be either larger or smaller than the usual – as an empirical matter they
are often found to be larger.
11
Wald and LM test:
F statistic robust to heteroskedasticity = wald statistic
LM statistic robust to heteroskedasticity
o Obtain the residuals 𝑢̃ from the restricted model
o Regress each of the independent variables excluded under the null on all of the included
variables; if there are q excluded variables, this leads to q sets of residuals (𝑟̃1 , 𝑟̃2 , … , 𝑟̃𝑞 )
o Find the product of each 𝑟̃𝑗 and 𝑢̃ (for all observations)
o Run the regression of 1 on 𝑟̃1 𝑢̃, 𝑟̃2 𝑢̃, … , 𝑟̃𝑞 𝑢̃ without an intercept. The heteroskedasticityrobust LM statistic is n-SSR1 where SSR1 is just the usualsum of squared residuals from the
final regression. Under H0 LM is distributed approximately as 𝜒𝑞2
Testing for hetereoskedasticity:
The tests have asymptotic justification under MLR1-4
We take the null hypothesis to be that assumption MLR5 is true.
- The Brusch-Pagan test
o Estimate the model by OLS and obtain the residuals. Compute the squared residuals 𝑢̂2
o Run the regression of the same model now with the squared residuals as the dependent
variable
o Form either F or LM statistic. F statistic = the test for overall significance of the regression.
LM statistic = 𝑛 ∙ 𝑅𝑢2̂2 ~𝜒𝑘2
- The White test
o Estimate the model by OLS as usual. Obtain the OLS residuals 𝑢̂ and fitted values 𝑦̂.
Compute the squared OLS residuals 𝑢̂2 and the squared fitted values 𝑦̂ 2
o Run the regression 𝑢̂2 = 𝛿0 + 𝛿1 𝑦̂ + 𝛿2 𝑦̂ 2 + 𝑒𝑟𝑟𝑜𝑟 Keep the r-squared from this regression
𝑅𝑢2̂2 .
o Form either F or LM statistic. F statistic = the test for overall significance of the regression.
LM statistic = 𝑛 ∙ 𝑅𝑢2̂2 ~𝜒22
Generalized least squares (GLS) estimators:
- Weighted Least Squares (WLS) estimators is more efficient than OLS estimators if we know the
form of the variance (as a function of explanatory variables)
Var(u|x)=σ2h(x) where h(x) is some function of the explanatory variables that determines the
heteroskedasticity. To get WLS estimates we divide the model by √ℎ𝑖
- Feasible generalized least squares (FGLS) estimators – here we model the function h and use the
data to estimate the unknown parameters in this model. This result in an estimate of each hi
denoted as ℎ̂𝑖 , that is we weight the model by 1/ℎ̂𝑖
o Run the regression of y on x1, x2, …, xk and obtain the residuals 𝑢̂
o Create Log(𝑢̂2 ) by first squaring the OLS residuals and then taking the natural log
12
o Run the regression of log(𝑢̂2 ) on x1, x2, …, xk and obtain the fitted values (𝑔̂)
o Exponentiate the fitted values: ℎ̂ = exp(𝑔̂)
o Estimate the model by WLS using 1/ ℎ̂
The squared residual for observation I gets weighted by 1/ℎ̂𝑖 . If instead we first transform all
variables and run OLS, each variable gets multiplied by 1/√ℎ̂𝑖 including the intercept.
FGLS estimators are biased but consistent and asymptotically more efficient than OLS.
If OLS and WLS produce statistically significant estimates that differ in sign or the difference in
magnitudes of the estimates is practically large, we should be suspicious. Typically this indicates
that one of the other Gauss-Markov assumptions is false. If MLR4 not is met then OLS and WLS
have different expected values and probability limits.
Chapter 9 – More on Specification and Data Issues
Endogenous explanatory variables: x is correlated with u
Exogenous explanatory variables: x is not correlated with u
RESET test for functional form misspecification:
The test builds on the fact that if the model satisfies MLR4 then no nonlinear function of the
independent variables (𝑦̂ 2 𝑎𝑛𝑑 𝑦̂ 3 ) should be significant when added to the equation.
In a RESET test polynomials in the OLS fitted values is added to the equation – normally squared
and cubed terms: 𝑦̂ = 𝛽̂0 + 𝛽̂1 𝑥1 + 𝛽̂2 𝑥2 + … + 𝛽̂𝑘 𝑥𝑘 + 𝛿1 𝑦̂ 2 + 𝛿2 𝑦̂ 3 + 𝑢
The null hypothesis is that the model is correctly specified. Thus RESET is the F statistic (F2,n-k-3) for
testing H0: 𝛿1 = 𝛿2 = 0 in the above auxiliary equation.
Test against nonnested alternatives:
Construct a comprehensive model that contains each model as a special case and then use F test
to test the restrictions that leads to each of the models.
Problem – a clear winner need not to emerge.
Using proxy variables for unobserved explanatory variables:
A proxy variable is something that is related to the unobserved variable that we would like to
control for in our analysis.
Assumptions needed for proxy variables to provide consistent estimators:
Model: 𝑦 = 𝛽0 + 𝛽1 𝑥1 + 𝛽2 𝑥2 + 𝛽3 𝑥3∗ + 𝑢 where 𝑥3∗ is unobserved
Proxy: x3
o The proxy should explain at least some of the variation in 𝑥3∗ . That is in the equation 𝑥3∗ =
𝛿0 + 𝛿3 𝑥3 + 𝑣3 a t-test of 𝛿3 should be significant.
o The error u is uncorrelated with 𝑥1 , 𝑥2 𝑎𝑛𝑑 𝑥3∗ . In addition u is uncorrelated with x3
o The variation not explained in the above mentioned equation (𝑣3 ) must not be correlated
with the other variables in the model (x1 and x2) or the proxy variable (x3)
13
Using lagged dependent variables as proxy variables:
We suspect one or more of the independent variables is correlated with an omitted variable, but
we have no idea how to obtain a proxy for that omitted variable.
Using lagged dependent variables in a cross-sectional equation increases the data requirements,
but it also provides a simple way to account for historical factors that cause current differences in
the dependent variable that are difficult to account in other ways.
Properties of OLS under measurement error:
Measurements error in the dependent variable – the usual assumption is that the measurement
error in y is statistically independent of each explanatory variable. If this is true, then the OLS
estimators are unbiased and consistent.
Measurement errors in an explanatory variable:
𝑥1∗ is not observed – instead we have a measure of it; call it x1
The measurement error in the population is: 𝑒1 = 𝑥1 − 𝑥1∗
We assume that u is uncorrelated with 𝑥1 𝑎𝑛𝑑 𝑥1∗ and E(e1)=0
What happens when simple replace 𝑥1∗ with 𝑥1 ? It depends on the assumptions we make about
the measurements errors
-
Cov(x1,e1) = 0 => cov(𝑥1∗ ,e1) ≠ 0
Here OLS estimation with x1 in place of 𝑥1∗ produces a consistent estimator of β1
-
Cov(x1,e1) ≠ 0 => cov(𝑥1∗ ,e1) = 0 Classical errors-in-variables (CEV) assumption
Here the variable we include in the model (x1) will be correlated with the error term (uβ1e1) Thus in the CEV case the OLS regression gives biased and inconsistent estimators.
We can determine the amount of inconsistency in simple OLS models:
𝑝𝑙𝑖𝑚(𝛽̂1 ) = 𝛽1 (
𝜎𝑥2∗
1
𝜎𝑥2∗ +𝜎𝑒21
)
1
𝑝𝑙𝑖𝑚(𝛽̂1) is always closer to zero than β1 when the CEV assumptions are met
-
If e1 is correlated with both 𝑥1∗ and x1 OLS is inconsistent
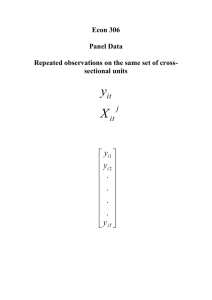
Chapter 13 – Polling Cross Sections across Time: Simple Panel Data Methods
Pooling independent cross sections across time:
One reason for using independently pooled cross sections is to increase the sample size. By polling
random samples drawn from the same population but at different points in time we can get more
precise estimators and test statistic with more power. Pooling is helpful only in this regard only
insofar as the relationship between the dependent variable and at least some of the independent
variables remains constant over time.
Typically we allow the intercept to differ across time at a minimum by including dummy variables
for the time periods (the earliest year is typically chosen as the base group).
14
Policy analysis with pooled cross section – natural experiment:
To control for systematic difference between the control and treatment groups, we need two
years of data, one before the policy change and one after the change. Thus, our sample is usefully
broken down into four groups: the control group before the change, the control group after the
change, the treatment group before the change and the treatment group after the change.
Call C the control group and T the treatment group, letting dT equal unity for those in the
treatment group T and zero otherwise. Then letting d2 denote a dummy variable for the second
time period, the equation of interest is:
𝑦 = 𝛽0 + 𝛿0 𝑑2 + 𝛽1 𝑑𝑇 + 𝛿1 𝑑2 ∙ 𝑑𝑇 + 𝑜𝑡ℎ𝑒𝑟 𝑓𝑎𝑐𝑡𝑜𝑟𝑠
𝛿1 measures the effect of the policy.
Without other factors in the regression 𝛿̂1 will be the difference-in-differences estimator:
𝛿̂1 = (𝑦̅2,𝑇 − 𝑦̅2,𝐶 ) − (𝑦̅1,𝑇 − 𝑦̅1,𝐶 )
Two-period panel data analysis:
In most applications, the main reason for collecting panel data is to allow for the unobserved
effect, ai, to be correlated with the explanatory variables.
𝑦𝑖𝑡 = 𝛽0 + 𝛿0 𝑑2𝑡 + 𝛽1 𝑥𝑖𝑡 + 𝑎𝑖 + 𝑢𝑖𝑡 , 𝑡 = 1,2.
t denotes the time period, d2t is a dummy variable that equals zero when t = 1 and one when t = 2
- it does not change across i, which is why it has no i subscript. ai captures all unobserved timeconstant factors that affect y. It is called an unobserved effect or fixed effect. The error u it is often
called the idiosyncratic- or time-varying error because it represents unobserved factors that
change over time and affect y.
Because ai is constant over time we can difference the data across the two years and hereby
‘difference away’ ai
𝑦𝑖2 = (𝛽0 + 𝛿0 ) + 𝛽1 𝑥𝑖2 + 𝑎𝑖 + 𝑢𝑖2 (𝑡 = 2)
𝑦𝑖1 = 𝛽0 + 𝛽1 𝑥𝑖1 + 𝑎𝑖 + 𝑢𝑖1 (𝑡 = 1)
(𝑦𝑖2 − 𝑦𝑖1 ) = 𝛿0 + 𝛽1 (𝑥𝑖2 − 𝑥𝑖1 ) + (𝑢𝑖2 − 𝑢𝑖1 ) 𝑜𝑟 ∆y = 𝛿0 + 𝛽1 ∆𝑥𝑖 + ∆𝑢𝑖
This is called the first-differenced equation and the estimators are called first-differenced
estimator
(The intercept is the change in the intercept from t = 1 to t = 2)
We can analysis the equation using the methods already developed, provided the key assumptions
are satisfied. The most important of these being that Δui is uncorrelated with Δxi
Another crucial condition is that Δxi must have som variation across i.
Assumptions for pooled OLS using first differences: FD1-5
FD1: For each I the model is:
𝑦𝑖𝑡 = 𝛽1 𝑥𝑖𝑡1 + ⋯ + 𝛽𝑘 𝑥𝑖𝑡𝑘 + 𝑎𝑖 + 𝑢𝑖𝑡 , 𝑡 = 1, … , 𝑇
FD2: We have a random sample from the cross section
15
FD3: Each explanatory variable change over time (for at least some i), and no perfect linear
relationships among the explanatory variables
FD4: For each t, the expected value of the idiosyncratic error given the explanatory variables in all
time periods and the unobserved effect is zero. E(uit|Xi, a1) = 0
Xi denote the explanatory variables for all time periods for cross-sectional observation I, thus it
contains xitj
Under FD1-4 the first difference estimators are unbiased
(FD4 is stronger than necessary – if E(Δuit|Xi) = 0 then FD estimators are unbiased)
FD5: The variance of the differenced errors, conditional on all explanatory variables, is constant
Var(Δuit|Xi) = σ2, t = 2, …, T
FD6: For all t ≠ s, the difference in the idiosyncratic errors are uncorrelated (conditional on all the
explanatory variables): Cov(Δuit , Δuis| Xi) = 0, t ≠ s
FD5 ensures that the differenced errors are homoskedastic. FD6 states that the differenced errors
serially uncorrelated, which means that the uit follow a random walk across time (something
explained in chapter 11!?).
Under FD1-6 the FD estimators of the βj is BLUE
Chapter 15 – Instrumental Variables Estimation and Two Stage Least Squares
IV – only one endogenous variable and one instrument
2SLS –one or multiple endogenous variables and more than one instrument
Assumptions for the instrumental variable z for x:
1. z is uncorrelated with u
2. z is correlated with the endogenous variable x
Cov(z,u) = 0
Cov(z,x) ≠ 0
IV estimators are consistent when the assumptions are met, but never unbiased why large
samples are preferred.
The instrumental variables (IV) estimator of 𝛽̂1 (simple regression):
𝑛
∑ (𝑧𝑖 −𝑧̅ )(𝑦𝑖 −𝑦̅)
𝐶𝑜𝑣(𝑧,𝑦)
𝛽̂1 =
= ∑𝑖=1
𝑛
𝐶𝑜𝑣(𝑧,𝑥)
𝑖=1(𝑧𝑖 −𝑧̅ )(𝑥𝑖 −𝑥̅ )
Homoskedasticity assumption (simple regression):
Needed for inference – now it is stated conditional on the instrumental variable z.
E(u2|z) = σ2
16
Asymptotic variance of 𝛽̂1 (simple regression):
𝜎̂ 2
̂
𝑣𝑎𝑟(𝛽1 ) =
2
𝑆𝑆𝑇𝑥 ∙ 𝑅𝑥,𝑧
The resulting standard error can be used for t-tests but F test are not valid since the R-squared
from IV estimation can be negative because SSR for IV can actually be larger than SST.
The IV variance is always larger than the OLS variance (since R-squared is always less than one and
this is the only thing that is different from the OLS formula – simple regression)
2
The more highly correlated z is with x, the closer 𝑅𝑥,𝑧
is to one, and the smaller is the variance of
2
the IV estimator. In the case that z = x, 𝑅𝑥,𝑧 = 1 and we get the OLS variance as expected
Weak correlation between z and x
Weak correlation between z and x can have serious consequences: the IV estimator can have a
large asymptotic bias even if z and u are only moderately correlated.
𝐶𝑜𝑟𝑟(𝑧,𝑢) 𝜎𝑢
𝑝𝑙𝑖𝑚𝛽̂1𝐼𝑉 = 𝛽1 +
∙
𝐶𝑜𝑟𝑟(𝑧,𝑥)
𝜎𝑥
Thus even if we focus only on consistency it is not necessarily better to use IV than OLS if the
correlation between z and x are smaller than between z and u.
Nothing prevents the explanatory variable or the IV from being binary variables.
IV estimation of the multiple regression model:
A minor additional assumption is that there are no perfect linear relationships among the
exogenous variables; this is analogous to the assumption of no perfect collinearity in the context
of OLS.
Reduced form equation: we writ an endogenous variable in terms of all exogenous variables.
- Used when testing the assumption cov(z,x) ≠ 0
2SLS – a single endogenous variable and more than one instrument:
E.g. with three instruments then: Cov(z1, z2, z3|u) = 0 and at least one of the instruments should be
correlated with the endogenous variable
Since each of z1, z2 and z3 is uncorrelated with u any linear combination is also uncorrelated with u.
To find the best IV we choose the linear combination that is most highly correlated with the
endogenous variables = the fitted values from regression the endogenous variable on all
exogenous variables in the model plus the instruments.
2SLS – multiple endogenous explanatory variables:
We need at least as many excluded exogenous variables as there are included endogenous
explanatory variables in the structural equation.
17
IV solution to errors-in-variables problems:
In the CEV case (x1 = x1*+ e1 and cov(x1,e1) ≠ 0) what we need is an IV for x1. Such an IV must be
correlated with x1, uncorrelated with u and uncorrelated with the measurement error e1.
One possibility is to obtain a second measurement of x1* - z1
- x1 and z1 both mismeasure x1*, but their measurements error are uncorrelated. Certainly x1 and
z1 are correlated through their dependence on x1*, so we can use z1 as an IV for x1
An alternative is to use other exogenous variables as IVs for a potentially mismeasured variable.
Testing for endogeneity of a single explanatory variable:
o Estimate the reduced from of the endogenous variable y2 by regressing it on all exogenous
variables (including those in the structural equation and the additional IVs) Obtain the
residuals 𝑣
̂2
o Add 𝑣
̂2 to the structural equation and (which includes y2) and test for significance of 𝑣
̂2
using an OLS regression. If the coefficient on 𝑣
̂2 is statistically different from zero, we
conclude that y2 is endogenous because (𝑣
̂2 and u are correlated). We might want to use a
heteroskedasticity-robust t-test.
Testing overidentification restrictions
Even in model with additional explanatory variables the second requirement (cov(z,x)≠ 0) can be
tested using a t-test (with just one instrument) or an F test (when there are multiple instruments).
In the context of the simple IV estimator we noted that the exogeneity requirement (cov(z,u) = 0)
cannot be tested. However, if we have more instrument than we need, we can effectively test
whether some of them are uncorrelated with the structural error.
The idea is that, if all instruments are exogenous, the 2SLS residuals should be uncorrelated with
the instruments, up to sampling error. The test is valid when the homoskedasticity assumption
holds
o Estimate the structural equation by 2SLS and obtain the residuals 𝑢
̂1
2
o Regress 𝑢
̂1 on all exogenous variables. Obtain R-squared, say 𝑅1
o Under the null hypothesis that all IVs are uncorrelated with u 1, 𝑛𝑅12 ~𝜒𝑞2 , where q is the
number of instrumental variables from outside the model minus the total number of
endogenous explanatory variables. If 𝑛𝑅12 exceeds the 5 % critical value in the 𝜒𝑞2
distribution, we reject H0 and conclude that at least some of the IVs are not exogenous.
18
Assumptions: 2SLS1-5
2SLS.1: Linear in parameters – The model in the population can be written as:
𝑦 = 𝛽0 + 𝛽1 𝑥1 + 𝛽2 𝑥2 + ⋯ + 𝛽𝑘 𝑥𝑘 + 𝑢
The instrumental variables are denoted as zj
2SLS.2: Random sample – we have a random sample on y, the xj and the zj
2SLS.3: i) There are no perfect linear relationships among the instrumental variables. ii) The rank
condition for identification holds.
2SLS. 4: The error term u has zero mean, and each IV is uncorrelated with u. (remember that any xj
that is uncorrelated with u also acts as an IV)
Under assumption 2SLS.1-4 the 2SLS estimator is consistent
2SLS. 5: Homoskedasticity - let z denote the collection of all instrumental variables. Then E(u2|z) =
σ2
Under assumption 2SLS.1-5 the 2SLS estimator is asymptotically efficient in the class of IV
estimators that uses linear combinations of the exogenous variables as instruments.
Instrumental variable estimation in matrix form:
The instrumental variable estimator in the just identified case:
𝛽̂𝐼𝑉𝐸 = (𝑍 ′ 𝑋)−1 𝑍′𝑌
The instrumental variable estimator of the predicted values with 2SLS: 𝑋̂ = 𝑍(𝑍 ′ 𝑍)−1 𝑍 ′ 𝑋 = 𝑃𝑍 𝑋
were 𝑃𝑍 = 𝑍(𝑍 ′ 𝑍)−1 𝑍 ′
- By this estimation the predicted X’s are exogenous and can be used in a second OLS step
The instrumental variable estimator in the over identified case:
−1
𝛽̂𝐼𝑉𝐸 = (𝑋̂ ′ 𝑋) 𝑋̂ ′ 𝑌 = (𝑋 ′ 𝑍(𝑍 ′ 𝑍)−1 𝑍 ′ 𝑋)−1 𝑋′𝑍(𝑍 ′ 𝑍)−1 𝑍 ′ 𝑌 = (𝑋′𝑃𝑍 𝑋)−1 𝑋′𝑃𝑍 𝑌
19