SDS -POLYACRYLAMIDE GEL ELECTROPHORESIS
advertisement

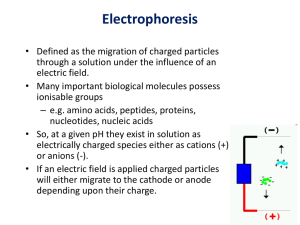
POLYACRYLAMIDE GEL ELECTROPHORESIS ( PAGE ) INTRODUCTION: Electrophoresis is the separation of charged molecules or ions in electric field. It is an analytical technique. Electrophoretic techniques are rapid and relatively sensitive requiring only micro grams of proteins. Electrophoresis of protein in polyacrylamide gel is advantageous than in any other gel and are carried out in buffers gels ( poly acrylamide native gels) as well as in sodium dodecyl sulphate (SDS) containing gels.[ denaturating conditions]. In the earlier method separation relies on both the charge and size of the protein. whereas in later, separation is based upon the size of the protein. Analysis of purity, determination of relative molecular weight of analytes are done in electrophoresis are accomplished in SDS – PAGE. The presence of isoenzyme is analsed in Native gel electrophoresis. SDS-PAGE: It is a technique used to separate proteins according to its size. Polyacrylamide gels are formed by polymerizing acrylamide with a cros s- linker (bisacrylamide) in the presence of a catalyst (sulphate radical) and chain initiator N ,N ,N’ ,N’ –tetramethylethylene diamine (TEMED). Solutions are normally degassed prior to polymerization , since oxygen inhibits polymerization. The porocity of the gel is determined by the relative proportion of acrylamide monomer to bisacrylamide[bis]. Gels are usually referred in terms of total percentage of acrylamide and bis present, and most protein separations are performed using gels in the range 7-15%. A low percentage of the gel is used to separate the high molecular weight proteins and vice -versa. 1. Separating gel buffer: 1.5 M Tris HCl, pH = 8.8. 2. Stacking gel buffer: 0.5 M Tris HCl, pH = 6.8. 3. Electrophoresis buffer: Dissolve 3.0 g of Tris base and the 14.4 grams of glycine in water and adjust the volume to 1 liter. The final pH should be 8.3. INSTRUMENTATION: The electrophoresis apparatus is set up with cathode buffer covering the gel in the negative electrode chamber, and anode buffer in the lower positive electrode chamber. Next, the denatured sample proteins are added to the wells one end of the gel with a micro pipette. Finally, the apparatus is hooked up to a power source under appropriate running conditions to separate the protein bands. An electric field is applied across the gel, causing the negatively-charged proteins to migrate across the gel towards the positive (+) electrode (anode). Depending on their size, each protein will move differently through the gel matrix: short proteins will more easily fit through the pores in the gel, while larger ones will have more difficulty (they encounter more resistance). After a set amount of time (usually a few hours- though this depends on the voltage applied across the gel; higher voltages run faster but tend to produce somewhat poorer resolution), the proteins will have differentially migrated based on their size; smaller proteins will have traveled farther down the gel, while larger ones will have remained closer to the point of origin. Therefore, proteins may be separated roughly according to size (and therefore, molecular weight), certain glycoproteins behave anomalously on SDS gels. Vertical Slab Mini Gel Electrophoresis: 1) Dimensions : 13cm(L) x 10cm(B) x 12cm(H) 2) No. Of Samples : 7 3) Buffer Volume : 250ml (including upper and lower tanks). 4) Complete unit consists of, Basic unit with Platinum Electrode Glass Plate notched & rectangular to run 8 x 7cm gel (2 Set) 7-well polished Teflon comb with thickness of1mmand 1.5mm (1No. each)(Well width = 7mm; Interspace = 3mm) Teflon Spacer with thickness of 1mm & 1.5mm (3 Nos. each) Gel Casting Stand (1 No.) Supporting Clamp (1 No.) Screws (8 Nos) and Metal Clips (2 Nos) and Lid & Power cord (1 No. each) PRINCIPLE: SDS is an anionic detergent which binds strongly and denatures proteins. The number of SDS molecules bound to a polypeptide chain is approximately half the number of amino acid residues in that chain. The protein - SDS complex carries net negative charges hence move towards the anode and the separation is based on the size of the protein. Electrophoresis is the study of the movement of charged molecules in an electric field. The generally used support medium is cellulose or thin gels made up of either polyacrylamide or agarose. Cellulose is used as support medium for low molecular weight biochemicals such as amino acid and carbohydrates whereas agarose and polyacrylamide gels are widely used for larger molecules like proteins. The general electrophoresis techniques cannot be used to measure the molecular weight of the biological molecules because the mobility of a substance in the gel is influenced by both charge and size. In order to overcome this, if the biological samples are treated so that they have a uniform charge, electrophoretic mobility then depends primarily on size. The molecular weight of protein maybe estimated if they are subjected to electrophoresis in the presence of a detergent sodium dodecyl sulfate (SDS) and a reducing agent mercaptoethanol. SDS disrupts the secondary, tertiary and quaternary structure of the protein to produce a linear polypeptidechain coated with negatively charged SDS molecules. 1.4 grams of SDS binds per gram of protein. Mercaptoethanol assists the protein denaturation by reducing all disulfide bonds. COMPONENTS: Chemical ingredients: Tris (tris (hydroxy methyl) aminomethane) (C4H11NO3; mW: 121.14): It has been used as a buffer because it is an innocuous substance to most proteins. Its pKa is 8.3 at 20 °C, making it a very satisfactory buffer in the pH range from roughly 7 to 9. Glycine (Amino Acetic Acid) (C2H5NO2; mW: 75.07): Glycine has been used as the source of trailing ion or slow ion because its pKa is 9.69 and mobility of glycinate are such that the effective mobility can be set at a value below that of the slowest known proteins of net negative charge in the pH range. The minimum pH of this range is approximately 8.0. Acrylamide (C3H5NO; mW: 71.08): It is a white crystalline powder. While dissolving in water, autopolymerization of acrylamide takes place. It is a slow spontaneous process by which acrylamide molecules join together by head on tail fashion. But in presence of free radicals generating system, acrylamide monomers are activated into a free-radical state. These activated monomers polymerise quickly and form long chain polymers. This kind of reaction is known as Vinyl addition polymerisation. A solution of these polymer chains becomes viscous but does not form a gel, because the chains simply slide over one another. Bisacrylamide(N,N'-Methylenebisacrylamide)(C7H10N2O2;mW:154.17): Bisacrylamide is the most frequently used cross linking agent for poly acrylamide gels. Chemically it is thought of having two-acrylamide molecules coupled head to head at their non-reactive ends. Sodium Dodecyl Sulfate (SDS)(C12H25NaO4S; mW: 288.38): SDS is the most common dissociating agent used to denature native proteins to individual polypeptides. When a protein mixture is heated to 100 °C in presence of SDS, the detergent wraps around the polypeptide backbone. It binds to polypeptides in a constant weight ratio of 1.4 g/g of polypeptide.Mobilities of these proteins will be a linear function of the logarithms of their molecular weights. Graph of logMW vs. Rf : The mobility (Rf) of a molecule in gel electrophoresis is determined by its free solution mobility, Y0 (= mobility in a gel of zero %) and the sieving action of the gel matrix. In practice the proportionality of log(MW) vs Rf holds true for most proteins, provided they are fully denatured, and provided the gel percentage has been chosen to match the molecular weight range of the sample. In fact, the actual plot of log(MW) vs Rf is sigmoidal (see figure below), because at high MW, the sieving affect of the matrix is so large that molecules are unable to penetrate the gel, while at low MW, the sieving effect is negligible, and proteins migrate almost at their free mobility, which in SDS is independent of MW. Although the overall graph of logMW vs. Rf is sigmoidal, it is nearly linear for a range of molecular weights depending on the percentage monomer of the gel. Given an appropriate selection of gel % and a protein which displays near-ideal behavior, molecular weights can be determined to within 5 - 10%. Molecular weights of non-ideal proteins can be determined by the use of Ferguson Plots, a technique which employs native protein electrophoresis. Ammonium persulfate (APS) (N2H8S2O8; mW: 228.2): APS is an initiator for gel formation. TEMED (N, N, N', N'-tetramethylethylenediamine) (C6H16N2; mW: 116.21): Chemical polymerisation of acrylamide gel is used for SDS-PAGE. It can be initiated by ammonium persulfate and the quaternary amine, N,N,N',N'-tetramethylethylenediamine (TEMED). The rate of polymerisation and the properties of the resulting gel depends on the concentration of APS and TEMED. Increasing the amount of APS and TEMED results in a decrease in the average polymer chain length, an increase in gel turbidity and a decrease in gel elasticity. Decreasing the amount of initiators shows the reverse effect. The lowest catalytic concentrations that will allow polymerisation in the optimal period of time should be used. APS and TEMED are used, approximately in equimolar concentrations in the range of 1 to 10 mM. Chemicals for processing and visualization: The following chemicals are used for processing of the gel and the protein samples visualized in it: Bromophenol blue(BPB) (3',3",5',5"tetrabromophenolsulfonphthalein)(C19H10Br4O5S; mW: 669.99): BPB is the universal marker dye. Proteins and nucleic acids are mostly colourless. When they are subjected to electrophoresis, it is important to stop the run before they run off the gel. BPB is the most commonly employed tracking dye, because it is viable in alkali and neutral pH, it is a small molecule, it is ionisable and it is negatively charged above pH 4.6 and hence moves towards the anode. Being a small molecule it moves ahead of most proteins and nucleic acids. As it reaches the anodic end of the electrophoresis medium electrophoresis is stopped. It can bind with proteins weakly and give blue colour. Glycerol (C3H8O3; mW: 92.09): It is a preservative and a weighing agent. Addition of glycerol (20-30 or 50%) is often recommended for the storage of enzymes. Glycerol maintains the protein solution at very low temperature, without freezing. It also helps to weigh down the sample into the wells without being spread while loading. Coomassie Brilliant Blue R-250 (CBB)(C45H44N3NaO7S2; mW: 825.97): CBB is the most popular protein stain. It is an anionic dye, which binds with proteins nonspecifically. The structure of CBB is predominantly non-polar. So is usually used (0.025%) in methanolic solution (40%) and acetic acid (7%). Proteins in the gel are fixed by acetic acid and simultaneously stained. The excess dye incorporated in the gel can be removed by destaining with the same solution but without the dye. The proteins are detected as blue bands on a clear background. As SDS is also anionic, it may interfere with staining process. Therefore, large volume of staining solution is recommended, approximately ten times the volume of the gel. n-Butanol (C4H10O; mW: 74.12): Water saturated butanol is used as an overlay solution on the resolving gel. Dithiothreitol (DTT; C4H10O2S2; mW: 154.25): DTT is a reducing agent used to disrupt disulfide bonds to ensure the protein is fully denatured before loading on the gel; ensuring the protein runs uniformly. Traditionally the toxic and less potent 2mercaptoethanol was used. METHEDOLOGY: 1 Thoroughly cleaned and dried glass plates and spacers were assembled properly.Held the assembly together with bulldog clips, in an upright position, 3% agar was then applied around the edges of the spacers to hold them in place and to seal the chamber between the glass plates. 2 Prepared a sufficient volume of separating gel; Tris-HCl : 1.5 M Tris HCl, pH = 8.8. Degassed for 3-5 minutes and then the following were added: Ammonium persulphate solution : 0.25 ml 10% SDS : 0.4 ml TEMED : 25 𝜇l 3 Mixed gently and carefully. Poured the gel solution in the chamber between the glass plates Layer the dist. water on top of the gel and left for 30 – 60 minutes for setting. 4 Prepared stacking gel (4%) by mixing the following solutions(total volume 10 ml): Tris- HCl : 0.5 M Tris HCl, pH = 6.8. Degassed and added the following constituents : Ammonium persulphate solution : 50 µl 10% SDS : 0.1 ml TEMED : 10 µl Removed the water from the top of the gel and washed with a little stacking gel solution. Poured the stacking gel mixture. Placed the comb in the stacking gel and allowed the gel to set. 5 After the stacking gel had polymerized, removed the comb without distorting the shape of the well. Carefully installed the gel after removing the clips, agar etc. in the electrophoresis apparatus. Filled it with electrode buffer and removed any trapped air bubbles at the bottom of the gel. Connected the cathode at the top and turned on the power briefly to check the electric supply. 6 Prepared samples for electrophoresis , following suitable extraction procedure. Adjusted the protein concentration in each sample using 5- strength sample buffer and water in such way that the amount of the protein is present per unit volume. 7 Heat sample solutions in boiling water for 2-3 min to ensure complete interaction between proteins and SDS. Cooled the sample solution and took the required volume in a micropipette and carefully injected it into the sample wells through the buffer. Marking the positions of the well on the glass plate with a marker pen and the presence of bromophenol blue in sample facilities easy loading of the samples. Similarly loaded a few wells with the std marker proteins in the sample buffer. 8 Turned on the current to 10-15 mA until the samples travel through the stacking gel. Then continue the run at 30 mA until the bromophenol blue reaches t he bottom of the gel. 9 10 After the run is complete, carefully remove the gel from between the plates and immersed in staining solution for at least 3hrs or overnight with uniform shaking.The separated proteins absorb the coomassie brilliant blue dye and thus can be visualized. To remove the dye that is not bount to the proteins, transfer the gel to a suitable container with at least 200-300 ml destaining solution and then is shook gently and continuously. Change the destainer and repeat the procedure till the background of the gel is colourless. SILVER STAINING: Silver staining was introduced by Kerenyi and Gallyas as a sensitive procedure to detect trace amounts of proteins in gels.It is used to visualize the fractionated polypeptides on gel by the silver staining technique. PRINCIPLE: The amino acids particularly aromatic aminoacids in the protein reduce silver nitrate and form complexes with metallic silver of yellowish-brown to brown colour. Silver staining is employed in the circumstances when the concentration of a protein is in minute amounts, which cannot be stained by coomassie brilliant blue R250 or amido black 10B dye.Silver staining has 100-fold greater sensitivity over dye staining. PROCEDURE: 1. 2. 3. 4. 5. 6. 7. 8. 9. After electrophoresis, the gel was transferred to a clean plastic container and washed in the washing solution with with slow shaking for 10 minutes. Discarded the wash solution and rinsed the gel with plenty of water for 2 minutes. Soaked the gel in sodium thiosulphate solution for 1-2 minutes. Washed the gel with water twice, each time 1-2 min. Drained the wash water. Soaked the gel in silver nitrate solution for 10 minutes with gentle soaking. Washed the gel as 4 mentioned above. Poured developer to the plastic container and shook the gel slowly, gently. The proteins reduce silver and the yellow to dark brown colour bands appear. When sufficient intensity of band developed, stopped the reaction by adding either citric acid or acetic acid solution. Recorded the protein banding pattern by photography. RESULT AND ANALYSIS: The fractionated proteins in the gel are observed as coloured bands of blue.As the proteins of minute quantities are stained faintly, destaining process should be stopped at appropriate stage to visualize as many bands as possible. The gel is then photographed and recorded. Silver stained SDS Polyacrylamide gels: Migration of proteins in SDS gels of varying acrylamide concentrations (%T). The migration of nine proteins ranging from 94 kDa to 14.4 kDa is shown. Stacking and unstacking occurs continuously in the gel, for every protein at a different gel concentration. The dotted line indicates the discontinuity at the Gly¯/Cl¯ moving boundary. Proteins between the fast leading electrolyte and the slow trailing electrolyte are not diluted by diffusion. SDS-PAGE Gel of Soluble Proteins: NATIVE GEL : Standard Curve Of Log: During SDS-PAGE, the intrinsic charges of the polypeptides are insignificant compared with the negative charges provided by the bound detergent, so that the SDS-polypeptide complexes have essentially identical charge densities and migrate in polyacrylamide gels of the correct porosity strictly according to polypeptide size and not charge. Since the distance migrated by a polypeptide is directly proportional to its size, the protein of interest can be identified in the gel profile if the molecular weight of its constituent polypeptides is known or, conversely, the molecular weight of individual polypeptides can be determined. To estimate the molecular weight of a sample polypeptide requires the investigator to construct a standard curve of log(polypeptide molecular weight) versus distance migrated for each of the standard molecular weight marker polypeptides run on the gel. This yields a straight line relationship. Knowing the distance migrated by the sample polypeptide, one can then read off its molecular weight from the curve. APPLICATIONS: SDS-PAGE has a number of uses, which includes; Determining protein size. Protein identification. Determining sample purity. Blotting applications.