docx - BeanBeetles.org
advertisement

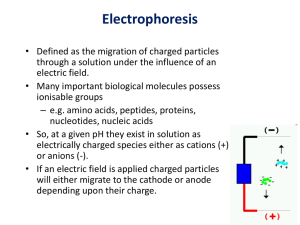
Protein expression in the life cycle of bean beetles (Callosobruchus maculatus) Instructor’s Notes Pre-laboratory Preparation You will need a number of cultures of bean beetles at various life stages. This will require cultivating cultures over an extended period of time so that the bean beetles are available at the time of the experiment. You will want the students to review the basics of protein synthesis as well as electrophoresis. Even though you will be doing protein electrophoresis, you can have students review DNA electrophoresis as tutorials on DNA electrophoresis are more readily available than protein electrophoresis. Experimental Design The students will be comparing proteins using protein electrophoresis. It is up to them to determine what proteins they want to compare. Some students may choose to compare males or females, larval stages versus adults or even between adults of different strains. The experimental protocol will basically remain the same in all instances. It may take some time for students to make the connection between DNA, proteins, and the appearance of organisms. It may be helpful to show them pictures of bean beetles at different life stages. Ideally you could show them pictures of a single individual at various life stages. Once they make the connection that the individual’s DNA has not changed but the appearance of specific proteins has through its life they can begin to think about what is causing the difference. Once they realize that proteins are playing a role in the differences they see they will begin to start formulating hypotheses. Following are the general guidelines for carrying out the lab. Be familiar with the protocol before attempting the lab in a classroom setting. There are three main parts: A. Extracting the protein and preparing it in a protein assay B. Determining the protein concentration of each of the samples – the purpose of this part of the protocol is to ensure that the concentrations of the samples are equal when placed in the gel for electrophoresis. If they are not equal the results will not be consistent and you cannot easily compare the proteins from different samples. However, if you have limited time you may choose to complete only the spot test and guesstimate the concentrations instead of completing the quantitation as well. You will still get results and should be able to see the differences. C. Vertical Gel SDS Page Electrophoresis A. Preparing the sample for protein assay Purpose: Extract the proteins from the bean beetles, denature the proteins and isolate the proteins from the other tissue present in the sample. Materials: o Solutions: sample buffer (without bromophenol blue(BPB)) o Eppendorf tubes, 2 per sample o Glass or metal rod for homogenizing the samples o Heat block Methods: 1. Label each of the sample buffer tubes. We recommend preparing more than one replicate per beetle type. For example, if you are looking at the difference between male and females, prepare 3-4 samples of males and the same for females. This way you will be able to compare within groups to ensure your results are consistent. 2. Take bean beetles in the corresponding tube and homogenize using a metal or glass rod. 3. Once fully homogenized leave the tube for 5 minutes to denature the proteins. 4. After 5 minutes transfer the liquid portion (not the tissue) to another blank eppendorf tube 5. Place it in a heat block at 95° C for 5 minutes. The samples are ready to be used to determine protein concentration. At this point, you can move onto part B to determine the concentration of the proteins in the various samples. The purpose of the next step is to ensure that the concentrations of the samples are equal when you place them in the gel. If they are not the same, the results will not be as consistent. However, if you do not have enough time to carry out Part B, you can complete just the first section, the spot test, before moving to the electrophoresis. In this case, you will have to guesstimate your protein concentrations. Generally, your results will still be visible, although they may not be as reliable. For more exact results, complete the spot test followed by the quantification. B. Determining protein concentration of samples: Coomassie stain protein assay Purpose: Determine the concentration of protein in each sample by comparing it to known protein standards. This assay can be used with protein samples in sample buffer without BPB. Protein standards (2, 4, 6, 7, and 10 mg Bovine Serum Albumin (BSA)/ml) can be prepared with either sample buffer or water and should be stored in the freezer. The procedure is to spot known amounts of the protein standards and your protein samples of unknown concentration onto whatman paper. Stain it with coomassie blue, then destain the paper. The amount of coomassie stain bound to your protein is proportional to the amount of protein in that spot. Adapted from Ghosh et al. 1988 Analytical Biochem. 169: 227 – 233. a. Spot test: Materials: o stain: 0.2% coomassie blue o destain: 10% acetic acid, 30% methanol in water o protein standards (2, 4, 6, 7, and 10 mg BSA/ml) o Whatman 3M paper o staining/destaining tray Methods: 1. Cut a clean piece of whatman 3M paper to fit in a small tray. Label it with a pencil to indicate the location of the samples and standards. 2. Spot 1 ul of each sample onto the paper and duplicate spots of each standard. Allow the paper to dry (15 - 30 minutes). You can speed this portion up by using a hairdryer to dry the paper. 3. Stain the paper at least 30 minutes. DO NOT shake. 4. Destain the paper, changing the destain when necessary. (30 minutes to an hour). 5. Remove from destain and allow to dry completely (you can speed up this drying process using a heat lamp or a hair dryer). 6. At this point you can either guesstimate your protein concentrations, or numerically quantitate them. b. Quantitation: Materials o Cuvettes o 2% SDS o Microfuge tubes o Hole punch o Colorimeter or spectrophotometer Methods 1. Cut the stained spots out of the paper with a hole punch and place them in microfuge tubes containing 1 ml 2% SDS. 2. Sonicate 1 hour or let sit overnight to elute coomassie from paper. 3. Read at 590nm. Preparing samples 1. 2. 3. 4. Label cuvettes and lids with sample labels. Label 5 cuvettes and lids with the different concentrations of BSA standard proteins Label a final cuvette and lid “Z” for Zero. Pipette 1 ml of the coomassie spot dot eluted BSA standards and your protein samples (your beetle protein extract) into the corresponding cuvette. Collecting data 1. Put 1 ml of 2% SDS into the Zero cuvette and wipe off the clear side with a tissue. 2. Place the Zero cuvette in the Colorimeter. Line up the clear sides with the arrow. Push the Zero button and remove cuvette when you see 0.00 3. Get one of the other cuvettes and wipe off the side with a tissue. 4. Place the cuvette in the Colorimeter. Line up the clear sides with the arrow. Push the Test button and record the absorbance values. 5. Continue this process with the other cuvettes until you have recorded the absorbance values for all of the protein standards and bean beetle samples. There are sample charts at the end of the lab. Determining protein concentration of samples 1. Open an Excel spread sheet containing a column for protein standard concentrations, and an additional column with their corresponding absorbances (as shown above). 2. Highlight the values, click on insert, and choose the scatter plot with no line function. 3. Go to chart layouts, choose trendline. In order to see the equation, go into trendline options and click “display equation on chart”. 4. Record the best fit line. 5. Determine protein sample concentrations of your samples by using displayed equation. Analyzing data and calculating dilutions for SDS-PAGE You will want to load an equal amount of protein for your protein gel. You may need to dilute your samples to be a standard 1 mg/ml of total protein. 1. For each sample, calculate how much of the sample to add to Laemmli Sample Buffer, with Bromophenol Blue to make 0.1 ml (= 100 l) volume. See the example at the end of the lab for help calculating. C. SDS Page electrophoresis1 Purpose: Separate the proteins onto a gel based on size and electrical charge. Vertical electrophoresis for proteins is a bit trickier and more complex than horizontal electrophoresis done with DNA. Materials: o Hot water bath or heat block o Vertical electrophoresis gel box o Vertical Gel o Protein ladder o Tris glycine/SDS buffer o Electrophoresis power supply o Coomasie blue stain o Destain o Lightbox Methods: 1. Take your samples out of the refrigerator/freezer. If samples are frozen, thaw. Place liquid protein samples into a 95° degree hot water bath or heat block for five minutes (CAUTION HERE!). 2. Inside the gel box is a piece called the gel cassette. Take the gel cassette out of the gel box. 3. On one side of the gel cassette is a buffer dam. If you are running one gel in the gel cassette leave the dam in place. If you are running two gels in the cassette, remove the dam. (DO NOT THROW THE DAM AWAY!) 4. Put on gloves and get a package that holds your gel. Turn the package over so you can see your gel. You should see the numbers written backwards like you see in the diagram to the right. THIS IS THE BACK OF THE GEL. When you see the writing correctly, that is the front of the gel. Open the package and remove the gel, try not to spill any of the liquid. Dispose of package in biohazard waste. Remove tape as directed on diagram. 5. Open the sides of the gel cassette. Place the gel in the gel cassette so you see the numbers written correctly from the outside of the gel cassette (This is critical so ask if you are unsure). Place the second gel in the same way or make sure the dam is placed correctly in the gel cassette. Close the sides of the gel cassette 1 Directions modified from work by: Nadja Anderson, PhD BIOTECH Project Director Molecular and Cellular Biology and BIO5 1007 E Lowell St University of Arizona Tucson, AZ 85721 Phone: 520-626-4664 Fax: 520-621-9903 email: nadja@bio5.org 6. Place the gel cassette into the gel box. Carefully remove the comb (The gel is very fragile), and load the gel with protein samples (15 uL). Don’t forget to record which sample is in which lane. Don’t forget to load a protein standard to monitor movement of proteins (10 uL). 7. Here is another critical step. Fill the inside of the gel cassette to the top with Tris glycine/SDS buffer. Outside the gel cassette, fill the gel box up to appropriate line (2 gel or 4 gel). 8. Plug the gel box into the power supply. Set the power supply to 200 volts and start the run. Watch the protein standard to know when the stop the gel. Lightest molecule of standard should about 1.5 centimeters from the bottom of the gel. 9. After the protein standard is down far enough in the gel, stop the power supply and remove the gel from the gel cassette. This should take about 35-45 minutes. Use a flathead screwdriver to crack open the gel casing and carefully (gel is thin and delicate) separate the two halves of the casing. The gel will stay on one side or the other of the casing and usually the bottom will curl up, that is a good place to gently pull gel up off the plate and place into the staining tray. BE CAREFUL OF ORIENTATION WHEN PLACING GEL IN STAINING TRAY! 10. Pour enough Coomassie blue stain to cover the gel. This is a powerful stain, be careful not to splash it on self or clothes. 11. Stain the gel, shaking gently for 30 minutes. After 30 minutes, recycle the stain by pouring it back into the bottle. 12. Replace the stain with destain (which will smell like vinegar) – just enough to cover the gel. Destain for about 30 minutes. Pour the blue destain down the sink (with the water running), gently rinse off the gel and pour fresh destain into the tray. Cover the tray with plastic wrap and place the tray into a large plastic bag. Destain overnight. 13. The next day, carefully dump the dirty destain down the sink (with the water running). Gently rinse the gel and place it on a light box for examination. Remember – The distance the protein band has traveled is related to the size of the protein. Also, how dark the band is tells how much of that protein there is – the darker the band, the more protein there is. Equipment and Supplies (for a class of 30 working in pairs) sample buffer (without BPB; see recipe below) 180+ Eppendorf tubes, 2 per sample 15 Glass or metal rod for homogenizing the samples Heat block and/or hot water bath Stain: 0.2% coomassie blue (see recipe below) Destain (see recipe below) protein standards (Sigma-Aldrich; Protein Standard 200 mg/ml # P5369-10ML: Dilute to make 2, 4, 6, 7, and 10 mg BSA/ml) whatman 3M paper 15 staining/destaining tray 100 Cuvettes 2% SDS 150-200 Microfuge tubes 15 Hole punchers Colorimeter or spectrophotometer (one per class; Montreal Biotech Inc.; CO 7500 Educational Colorimeter) Vertical electrophoresis cell (bio-rad; Mini-PROTEAN® Tetra Cell for Mini Precast Gels #1658004: This version holds 4 gels. There are also versions that hold two gels.) 7-8 Vertical Gels (bio-rad; 12% Mini-PROTEAN® TGX™ Precast Gel # 456-1043S: This comes with 10 precast gels) Protein ladder (bio-rad; Polypeptide SDS-PAGE Standards # 161-3026) Tris glycine/SDS buffer (enough to fill 2 gel boxes; see recipe below) Electrophoresis power supply 7-8 Lightbox (or share light boxes between groups) Example equation for quantification: Example: if the measured concentration of your sample was 3 mg/ml, use the following formula to make a final volume of 0.1 ml of 1 mg/ml protein: (final concentration) X (final volume) = (sample concentration ) X (sample volume) (final concentration) X (final volume) --------------------------------------------- = sample volume Sample concentration (1 mg/ml) (0.1 ml) = (3 mg/ml) (x) (1 mg/ml) (0.1 ml) ----------------------- = (x) (3 mg/ml) x = 0.0333 ml = 33.3 l Volume of Laemmli Sample Buffer needed = (100 l – sample volume) Volume of Laemmli Sample Buffer needed = (100l – 33.3l) = 66.7l Laemmli Sample Buffer for a final concentration of 1 mg/ml. Recipes and Background Information 5X stock of Tris-Glycine buffer Combine 15.1 g Tris base and 94 g glycine with water to make a total volume of 1 liter. Dilute this 1:4 with water (i.e. 100 ml stock with 400 ml water to make a total 500 ml of 1X Tris-Glycine buffer). Store in a sealed bottle at room temperature indefinitely. 3% agarose in Tris-Glycine buffer In a 250 ml Pyrex bottle, combine 3.75 g agarose with 125 ml Tris-Glycine buffer - DO NOT USE TRISGLYCINE-SDS BUFFER FOR THIS! You'll end up with a giant bubbly mess. Microwave uncovered for 1 minute at a time until agarose is dissolved, being careful not to let the agarose boil over on the microwave or on you! Store loosely covered at room temperature until solidified, then tightly cover to store indefinitely. Be sure to remove cover before you microwave to dissolve the agarose again. 5X stock of Tris-Glycine-SDS buffer Combine 15.1 g Tris base, 94 g glycine, and 50 ml 10% SDS (5 g SDS with 45 ml water) with water to make a total volume of 1 liter. Dilute this 1:4 with water (i.e. 100 ml stock with 400 ml water to make a total 500 ml of Tris-Glycine-SDS buffer). Store in a sealed bottle at room temperature indefinitely. Coomassie blue stain For each liter of stain, combine 450 ml water, 2.5 g Coomassie blue, 450 ml methanol, and 100 ml glacial acetic. Store in a sealed bottle at room temperature indefinitely. Destain For each liter of destain, combine 600 ml water, 300 ml methanol, and 100 ml glacial acetic acid. Store in a sealed bottle at room temperature indefinitely. 1.0 M Tris-Cl (pH 6.8) Combine 60.5 g Tris base with about 350 ml water. Add enough HCl to give the solution a pH of 6.8. Add enough water to make the total volume of the solution 500 ml (0.5 liter). Store in a sealed bottle at room temperature indefinitely. Sample buffer To make 100 ml sample buffer, combine 10 ml 1.0 M Tris-Cl (pH 6.8), 20 ml 20% SDS, 0.1 g bromophenol blue, 20 ml glycerol, and water to make a total volume of 100 ml. This is actually a 2X recipe, but the sample buffer is used at 2X, not diluted. Dispense in 0.5 ml aliquots for use by the students. Store at room temperature indefinitely. This study was written by T. Ashraf and E. Priddis, 2013 (www.beanbeetles.org).