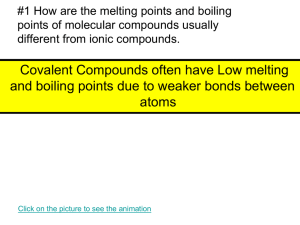
Molecular Modeling
advertisement

CHEMISTRY 51 MOLECULAR MODELING INTRODUCTION. This set of laboratory exercises is designed to acquaint you with selected but instructive techniques of molecular modeling. Computational chemistry has evolved to the point that a good calculation can be the full equivalent of an experiment. The contemporary chemical literature makes frequent reference to in silico chemistry that is chemistry performed on a computer. The following exercises are designed as a series of experiments in which you will use computational methods to answer chemical questions. Computational chemistry covers a broad range of methods and theory and no attempt will be made to provide an exhaustive coverage of the field. However, we shall address problems that most scientists identify as central to the evolving discipline of molecular modeling. This set of exercises will primarily focus on applications that use techniques from quantum mechanics. The central result is the Schrödinger wave equation: H = E where the wave function is a mathematical representation of the system, i.e. a virtual molecule, H represents the measurement of the energy, and E is the experimental value of the energy. Physicists and physical chemists assert that the principles of quantum mechanics are sufficient for the understanding of all chemical phenomena. When Dirac made this bold claim in the early 1920’s, there was no means of solving the Schrödinger equation for any non-trivial chemical system. Although the theory was complete in principle, it was useless in practice as the equation could not be solved. Nearly a century of intense theoretical effort and the development of computers have solved this problem and it is now possible to obtain chemically useful results from a numerical solution of the Schrödinger equation. Several programs have been developed to this end. Gaussian is the leading program for serious quantum chemistry but it is particularly difficult to use. We shall be using Spartan ‘02, a powerful yet user-friendly program. The College has a site license for Spartan that supports up to 15 simultaneous users. Although we shall be using it in one of the department’s computer labs, you can also run the software on any computer in a campus computer lab that is running under Windows 2000. PLAN OF ATTACK. We shall be using the computers in Seaver North 113 and will take advantage of the physical arrangement of the hardware. Two computers are located at each of the 6 tables in the room. Accordingly, each problem involves calculations and analysis for two molecules. You will work in pairs. In each pair, one student will use his/her computer to study one of the molecules while the partner will work with the second molecule on her/his computer. Each group will compare results of the calculations. This format is well suited to the practice of molecular modeling where informative results are usually obtained when some parameter is changed. 1 This document will not outline every keystroke. Rather, the instructor will demonstrate the use of the program and will be available to handle problems as they develop. If you wish to familiarize yourself with the program before lab, consult the tutorial, Spartan ’02 Tutorial and User’s Guide. A copy is on reserve in the Seeley G. Mudd Library. An electronic version of the tutorial is available through the Help function of the program. Spartan is relatively easy to use and you will be able to fly solo after the lab session. There are more exercises in this document than we can cover during the lab period. We encourage you to pursue them and to use Spartan in cases where the application of molecular modeling is instructive. Those of you who take Organic Chemistry next year will make additional use of Spartan. When you arrive in lab (Seaver North 113 for this exercise), select a computer and log on. Click on the Spartan ikon to start Spartan. We encourage you to save all the work in your own space since the disk on a public computer is cleaned up when the user logs off. You are probably familiar with this routine from your use of Excel. MODELING EXPERIMENTS 1) Validation of Valence-Bond Models of Acids and Conjugate Bases In class, we used valence-bond theory in its Lewis electron-dot form to discuss bonding theory. However, chemical bonds are a useful human construction developed to understand the properties of molecules and electrons in molecules. That is, electrons, not bonds, are real. In this experiment, you will compare the predictions from a VB treatment for formic acid (HCOOH) and its conjugate base, the formate anion (HCOO-) with the results of quantum mechanical calculations, i.e. virtual experiments. Before you come to lab, draw Lewis electron dot structures of formic acid and the formate anion. One member of each group should select formic acid and the other formate anion. Complete the following steps. a) Draw the structure of the molecule by selecting Carboxylic Acid from the Groups menu. The structure is complete in the case of formic acid. The molecular editor will automatically add hydrogens to the yellow dangling bonds. In the case of the formate anion, click on Delete in the Geometry menu and delete the dangling bond connected to the carboxyl oxygen by clicking on it. Then no hydrogen atom will be added to this oxygen when you leave the molecular editor. b) Set up a calculation. Specify the Task in the Calculate boxes as Equilibrium Geometry at the Ground state. The method should be Hartree-Fock with a 3-21G* basis functions. Also click on the box marked E. Sol. This option will produce an estimate of the species’ electronic energy in water. In the case of the formate anion, one more change is necessary. Set the Total Charge box to read anion. Then click on the Submit button and enter a file name and path as prompted. c) When the calculations have been completed, measure the bond angles and bond lengths of the two species. Discuss the comparative results. The 2 values of the energy for each species in vacuo and in aqua are obtained by accessing the Output submenu of the Results menu. Does hydration stabilize or destabilize these species? Will solvation increase the extent of dissociation? 2) Strength of Species with Lone Pairs as Ligands and Bases G. N. Lewis defined a base as an electron donor. Bases can also function as ligands as the basis for a coordinate-covalent bond between a ligand and a transition metal species is the sharing of an electron pair between the two. Accordingly, ammonia which has one lone pair acts as both a base and a ligand. Phosphorus is in the same family as nitrogen and phosphine, PH3, should also have the same electronic properties. However, this qualitative argument does not allow one to predict the relative reactivity of these two species. The pKb’s of ammonia and phosphine are 4.75 and 25.67, respectively; but both species are good ligands. The goal of this is exercise is to understand these striking differences with the aid of molecular modeling. One student should choose ammonia and the other phosphine. Perform the following steps: a) Draw the structure of your molecule. b) Set up a calculation. In the Calculate windows, select Equilibrium Geometry with Hartree-Fock and a 3-21G* basis set. Also click on Elect. Charges. Click on the Submit button and provide a path and file name as prompted. c) When the calculation is finished, measure the bond angle in your compound. Is the value consistent with bonding theory? Also, use the Output function to read the charges on the atoms. The key strokes of this and later steps will be demonstrated by the instructor with water as an example. Are your results consistent with your notion of electronegativity? d) An electron density surface can be an informative graphical presentation of information. This is a surface with the property that the electrons in the molecule will be found on or inside the surface a certain fraction of the time. This fraction is given by one minus the isovalue parameter. Hence with the default isovalue = 0.002, the electrons will be found on or inside the surface with 99.8% probability. Brønsted defined a base as a proton acceptor. A potential surface can show the electrical attraction between a species and a proton. A potential surface is a surface on which the electrical potential has a fixed value. Generate these surfaces using the Setup menu and the Surfaces submenu. To prepare for the generation of the surfaces, click on the Add button and select first density from the menu of surfaces with potential as the property and then potential from the menu of surfaces with no property. To actually generate the surfaces selected above, click on the Submit submenu in the Setup menu. When the process is completed, the unfilled square to the left of the 3 name of the surface will turn yellow. To display the surface, click on the square; a red check will appear. To undisplay, click on the square again. e) Use the graphical and quantitative results of the modeling calculations to discuss the chemistry of ammonia and phosphine. 3) The Molecular Structure of Sulfur Tetrafluoride (SF4) We have used the VSEPR model, an extension of the valence-bond (VB) model, to predict the three-dimensional structure of molecules. In its simplest form, the model asserts that the arrangement of substituents and lone pairs attached to a central atom is determined by repulsion between electrons. For example, in the case of BF3, three groups of charge belonging to the three F atoms distribute themselves to form an equilateral triangle. The well-known trigonal planar structure results. In many cases, lone pairs of electrons compete with bonding pairs. The text book presents a hand waving argument that lone pairs occupy more space than bonding pairs but the argument is by no means convincing. In cases such as this, the VSEPR model provides a set of alternatives and a plausibility argument for a preferred structure. An experiment is required to provide a convincing case for or against the hypothesis. One experimental approach would be the measurement of the microwave spectrum of the compound. This spectrum yields the moments of inertia of the molecule. In the case of small, polar molecules, this information is sufficient for the determination of its three-dimensional structure. Molecular modeling provides an alternate experimental approach. Once calculates the electronic energy for each molecular structure. The structure that yields the lowest energy is the preferred form. Before you come to lab, use the VB/VSEPR model to determine reasonable hypotheses for the three-dimensional structure of SF4. In lab, each group of two should select two candidate structures from the list and each member of the group will choose a different structure. Complete the following steps for each structure: a) Draw the structure. You will need to use the expert mode. Don't forget to delete any unused dangling bonds. You don't draw in the lone pairs. Their number and location are determined by quantum mechanics and the program. b) Set up a calculation. Specify the Task in the Calculate boxes as Equilibrium Geometry at the Ground state. The method should be Hartree-Fock with a 3-21G* basis functions. Also click on the Freq. box. Accept the default values for the other options. Click the Submit button and provide a path and file name as prompted. The program will accept the structure that you input as the starting point of a routine in which the electronic energy is calculated and the structure is adjusted to minimize this energy. This procedure will determine the structure closest to the starting point that has a minimum energy. This is called the local minimum. c) When the calculation has completed, determine the energy using Display menu and the Output submenu. Record the electronic energy in atomic units. Also measure the bond angles and bond lengths of any stable species. and check using the Display menu and the Vibrations submenu the list of 4 vibrational frequencies calculated for your structure. All frequencies of a stable molecule must be real numbers. An activated complex, a species that we shall encounter in chemical kinetics, has one imaginary frequency. d) Compare the results of the two calculations. They should allow us to determine the most stable isomer or conformer of SF4. Does SF4 have more than one stable form? We shall discuss the results in lab. Do the calculations refute or support your hypothesis? Interpret the values of the bond lengths and angles. 4) Carbon Monoxide and Cyanide as Ligands. Most people know that cyanide and carbon monoxide are potent poisons but are probably unaware of the chemical basis for this property. We have noted in class that these species are poisons as they are strong ligands and bind to transition metal species such as iron(II). We did not address the orientation of the ligand when it forms a coordinate covalent bond. Is it M-CX or M-XC where X is N or O? The preference for one orientation or isomer will be explored in this exercise. To answer the question, we should display the molecular orbital with the highest energy, the so-called Highest Occupied Molecular Orbital or HOMO for short. Species that are electron deficient attack electrons in this orbital and hence of all the molecular orbitals in the molecule, the HOMO is most indicative of the species chemistry. Fukui shared the Nobel Prize with Hoffman and Woodward for developing the study of these “frontier” orbitals. Similarly, in examining the chemistry in which a reagent donates charge to a molecule under study, the most instructive orbital for this chemistry, i.e. the relevant frontier orbital, is the Lowest Unoccupied Molecular Orbital or LUMO. As in the previous exercises, one member of the pair should choose carbon monoxide and the other cyanide. As before, complete the following steps: a) Draw the structure of your compound. Use a triple bond between the two atoms in the ligand. In the case of cyanide delete the dangling bond or a hydrogen atom will be appended when you leave the molecular editor. b) As before, set up an energy calculation. Use Hartree-Fock and a 3-21G* basis set. Be sure to provide the charge if your ligand is an ion. Submit the job as before. c) When the calculation is completed, draw the HOMO. This is accessed via Setup and Surfaces. You used this utility before to draw the electron density and potential surfaces. This same approach is used in this case but HOMO is selected as the surface. d) Examine the HOMO. Which end of the ligand will be the preferential site of attack? mol_model_lab.doc, 27 June 2001, WES 5