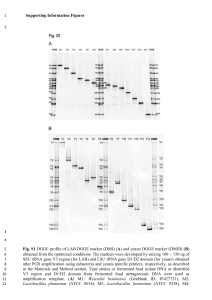
Finding curly leaf
advertisement

GMO Bioinformatics Exercise: Finding DNA Using BLAST (‘Electronic PCR’) Instructions and Worksheet Save this document to your desktop and complete it on your computer! You may also email it to yourself for save-keeping. Task: Determine the nature of the DNA sequences used to A) identify transgenic plants and to B) identify DNA in foods (positive control). A) Primer Set A for transgenic plants: 5'-CCGACAGTGGTCCCAAAGATGGAC-3' (Forward Primer), 5'-ATATAGAGGAAGGGTCTTGCGAAGG-3' (Reverse Primer). B) Primer Set B to probe for the presence of DNA (positive control): 5'-GGGATCCACTTCATGCTTTCGTCC-3' (Forward Primer), 5'-GGGAACCACATCACCACGGTACAT-3' (Reverse Primer). 1) Set up the experiment a. Open the website of the National Institute for Biotechnology Information (NCBI) at http://www.ncbi.nlm.nih.gov/). b. Find and click the link to ‘BLAST’. c. Find and click the link to a ‘Nucleotide-nucleotide BLAST (blastn)’. d. Highlight and copy the sequences for Primer Set A above and paste it into the search window; eliminate any non-nucleotide characters from the sequence. e. Click . f. Click . 2) Analyze the search results a. How many search hits were identified? b. Did the search turn up any significant results as opposed to hits that came up just by chance? (In order to answer this question locate the column labeled ‘E-values’. Low Evalues denote higher significance, E-values of 0.1 and higher indicate insignificant hits. (Example: 3e-04 denotes an E-value of 3x10-04 or 0.0003, indicating a significant hit. Bitscores are usually no good indicators for the significance of a hit. Learn more about Evalues in the ‘BLAST FAQs’-section of the result page.) 3) Analyze a search hit a. Each hit represents a DNA sequence that is saved in a unique datasheet in a database called GenBank. Each GenBank entry has an accession number that, usually begins with ‘>gi|’. Click the accession number for any of the hits listed. b. Read the data sheet and determine the general nature of the hit. (How many nucleotides is it long? When was it entered into GenBank? Who entered it? What kind of sequence is displayed – genomic or plasmid DNA? What organism is it from?) c. Click the browser’s ‘Back’-button and go back to the listing of search results. d. Scroll to the end of the search result list until you arrive at the section that shows detailed alignments of DNA sequences the query sequences (‘Query’) with the hits. e. Identify a hit that contains alignments of hit sequences (‘Sbjct’) with both primers and record its Accession number. f. Record the highest and lowest coordinates of the positions where ‘Query’ (primers) and ‘Sbjct’ sequences match. Lowest coordinate Highest coordinate Forward Primer Reverse Primer g. Access the GenBank data sheet for this hit and, using the coordinates from the table above, identify in the DNA sequence the regions that match the primers. h. Determine the distance of the two matching regions. Then, determine the length of the DNA fragment that is bordered by the two primer sequences and enter it into the box. Fragment length (bp) i. How does this DNA fragment correspond to the Fragment amplified in the real PCR reaction performed on plant and food DNA in the lab? 4) Nature and function of the DNA identified by the BLAST search a. Many DNA sequences in GenBank are annotated, providing details such as the location of genes and promoters. Find the annotation for the hit you examined under 3) and determine whether it contains any annotation for the region that matches the two primers. b. What is the function and origin of this DNA region? (A search engine such as ‘Google’ may prove helpful to determine the ultimate function and source of the DNA element the primers match with). 5) Repeat steps 1) through 4) for Primer Set B. What is the gene used as positive control?