Supplementary information (Nakagawa et al “Structure and
advertisement

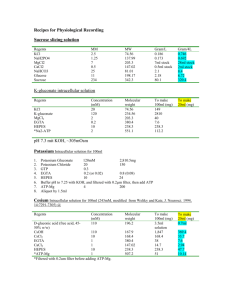
Supplementary Information and Figure Legends (Nakagawa et al “Structure and different conformational states of native AMPA receptor complexes”) Purification of AMPA-R and Fab labeling 150 - 300 adult rat brains were homogenized in 20 mM HEPES, pH 7.4, 320 mM sucrose, 5 mM EDTA, 5 mM EGTA, 30 M NBQX supplemented with protease inhibitors (1 mM PMSF, 10 g/ml aprotinin, 10 g/ml leupetin, 1 g/ml pepstatin, and 500 M benzamidine). Supernatant obtained by centrifuging the homogenate at 3,000 g for 15 min was further spun at 38,400 g for 15 min to obtain a membrane pellet (P2 fraction). P2 was resuspended in SB1 (20 mM HEPES, pH 7.4, 1 M KI, 5 mM EDTA, 5 mM EGTA, and 30 M NBQX) and membranes were collected by centrifugation. Membranes were further washed with SB2 (20 mM HEPES, pH 7.4, 4 M urea, 5 mM EDTA, 5 mM EGTA, 30 M NBQX) to remove associated proteins and collected again by centrifugation. Finally, membranes were solublized in RB (20 mM HEPES, pH 7.4, 100 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1 % CHAPS, 30 M NBQX, with protease inhibitors) for 30 min with gentle stirring and ultracentrifuged at 100,000 g to remove insoluble material. The final supernatant was applied to an anti-GluR2 C-terminus antibody affinity column (0.5 ml bed volume, antibody concentration 2 mg/ml). After washing the column with 3 ml of sample buffer, GluR2 containing AMPA-Rs were eluted with 20 mM HEPES, pH 7.4, 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1% CHAPS, 30 M NBQX, 0.5 mg/ml GluR2 C-terminal epitope peptide (GYNVYGIESVKI). The proteins were further purified on a Superdex 200 gel filtration column (Pharmacia) in 20 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 0.3 mM ZnSO4, 1% CHAPS. Zinc was added in the final buffer because it is found in the dimer interface of the ligand binding domain crystals1. When indicated 1% CHAPS was substituted by 0.1% dodecyl maltoside or 0.6% decyl maltoside. Western blotting (Supplementary Fig. 1b) and mass spectrometry analysis (not shown) confirmed the presence of AMPA-R subunits in the ~100 kDa band. The proteins GRIP 2,3 and SAP97 4, which bind to the cytoplasmic tails of AMPA-Rs, were absent in the final preparation (Supplementary Fig. 1b). Polyclonal antibodies against the following peptides were raised in rabbits and affinity-purified with the antigen peptides. C-terminus of GluR1 (SHSSGMPLGATGL), C-terminus of GluR2 (EGYNVYGIESVKI), NTD of GluR1 (RTSDSRDHTRVDWKR), pan-TARP#1 (CIQKDSKDSLHANTANRRTTPV), and pan-TARP#2 (CPEDADYEADTAEYFLR). Fab fragments were purified using the Immunopure IgG1 F(ab’) and the F(ab’)2 Fab purification kit (Pierce) followed by gel filtration on a Superdex 200 column (Pharmacia). Labelling was performed by incubating AMPA-Rs with Fab fragments at a molar ratio of 1:4 to 1:8 overnight at 4oC in 20 mM HEPES, pH 7.4, 100 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1% CHAPS. Image processing Electron micrographs were digitized with a SCAI scanner (Zeiss) using a step size of 7 m. 3 x 3 pixels were averaged yielding a pixel size on the specimen level of 4.04 Å for the negatively stained and 5 Å for the cryo-negatively stained specimens. Image processing was performed with the SPIDER software package 5. For conventional negatively stained specimens, particles were selected interactively using WEB, the display program associated with SPIDER, and windowed into small images of 100 x 100 pixels. Projection averages were calculated over 10 cycles of K-means classification and multireference alignment specifying 100 output classes. For 3D reconstructions of the cryo-negatively stained preparations, particle pairs were interactively selected from a total of 373 image pairs of 50o/0o tilted specimens and windowed into small images of 100 x 100 pixels. Table 1 summarizes the number of particles used in this study. The particles from the images of the untilted specimens were used for classification into 100 classes as before. The images of the tilted specimens in each class were used to calculate initial 3D reconstructions of individual classes by backprojection, backprojection refinement, and angular refinement. The final volume obtained by angular refinement with SPIDER was used as input model for FREALIGN, which was used for further refinement of the orientational parameters of the individual particles and to correct for the contrast transfer function of each particle image according to its defocus value 6. The correct defocus value for each particle image was deduced from the position of each particle in the image and the tilt angles and defocus values of the images, which were determined with CTFTILT 7. Electron micrographs showing a high degree of astigmatism were rejected. Particles selected from both the images of the tilted and the untilted specimens were used for the refinement with FREALIGN. The resolution of the final 3D reconstructions were estimated using Fourier shell correlation (FSC) and ranged from 42 to 46 Å using the FSC = 0.5 criterion or from 31 to 33 Å using the FSC = 0.142 criterion proposed by Rosenthal and Henderson 8. Supplementary Figure 1. Purification of AMPA-Rs. a. Fractions eluting from the anti-GluR2 immuno-affinity column (“AMPA crude”) and the subsequent gel filtration (Superdex 200) column (fractions 13 – 21) were resolved by SDS-PAGE (4 – 15 % gradient gel) and visualized by silver staining. The flow rate was 0.5 ml/min and 0.5 ml fractions were collected. Fraction 17 was used for EM. b. Fractions shown in (a) were immunoblotted for the proteins indicated on the left. Brain extract (3,000 g centrifugation supernatant of brain homogenate) was used as positive controls for immunoblots against GRIP and SAP97. Supplementary Figure 2. Plot of the angle distributions and comparison of reprojections from 3D models with the corresponding raw particle images. a. Distribution of the angles (phi and theta) assigned by FREALIGN to the particle images used for the 3D reconstruction shown at the left bottom of each plot. Theta is zero at the center and 90o at the outer edge of the circle. Phi is zero at the position indicated in the figure and increases radially in counter-clockwise direction. The distribution is consistent with the random conical tilt reconstruction approach, where the majority of particles are distributed around phi = 50o and the images from the untilted specimens (phi = 0o) are concentrated in the center of the plot. b. Representative side-by-side comparisons of raw particle images (left panels) with the corresponding reprojections from the final 3D reconstructions (right panels). Supplementary Figure 3. 3D density map of AMPA-R in the type I conformation filtered to 42 Å (FSC = 0.5 criterion) or 31 Å (FSC = 0.142 criterion). The difference between density maps filtered to a resolution of 31 Å or 42 Å is virtually undetectable for a particle the size of AMPA-R and makes no differencce for the placement of crystal structures into the density map. We therefore filtered the density maps shown in Fig. 2 to the more conservative resolution value of 42 Å. Supplementary Figure 4. ClustalW alignment of mGluR1 (extracellular domain), LIVBP, and GluR2-NTD. The amino acid residues present in the extracellular domain of mGluR1 but not in GluR2 are highlighted in yellow. These residues were removed from the crystal structure that was placed into the EM density map in Fig. 2. Supplementaty Figure 5. Class averages of particles obtained under different drug treatments. Negative stain EM images of particles from the same preparation, left untreated (a), treated with 1 mM glutamate (b), or with 1 mM glutamate plus 330 M CTZ (c). Particles were classified into 100 classes by multivariate classification (K-means classification) and multi-reference alignment. The orientations of each class average image are not the same. Note the lack of clear Type I particles in the glutamate treated class averages (b). To quantify the change of the distribution of the conformation we assigned each class average as “unclassified” (O), “Type I” (I), or “Type II” (II). The number of particles contained in each class average is indicated in the bottom right of each box. Supplementary Table 1. Peptides and corresponding proteins identified by LC/MS/MS tandem mass spectrometry analysis of the bands indicated by asterisks in Fig. 3a. References 1. 2. 3. 4. 5. Sun, Y. et al. Mechanism of glutamate receptor desensitization. Nature 417, 24553 (2002). Dong, H. et al. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature 386, 279-84 (1997). Wyszynski, M. et al. Association of AMPA receptors with a subset of glutamate receptor-interacting protein in vivo. J Neurosci 19, 6528-37 (1999). Leonard, A. S., Davare, M. A., Horne, M. C., Garner, C. C. & Hell, J. W. SAP97 is associated with the alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR1 subunit. J Biol Chem 273, 19518-24 (1998). Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116, 190-9 (1996). 6. 7. 8. Grigorieff, N. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 A in ice. J Mol Biol 277, 1033-46 (1998). Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142, 334-47 (2003). Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333, 721-45 (2003).