Word file (1.33 MB )
advertisement

Supplementary Material
Materials and Methods
1.
Methods of Logical Inference
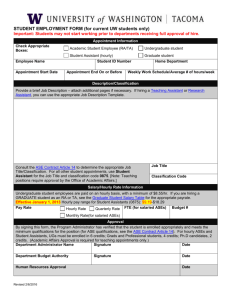
A comparison of deduction and abduction as methods of logical inference.
Deduction
Rule
Fact
∴
If a cell grows on minimal medium, then it can synthesise
tryptophan.
Cell cannot synthesise tryptophan
Cell cannot grow on minimal medium.
Given the rule P Q, and the fact Q, infer the fact P (deduction - modus tollens)
Abduction
Rule
Fact
∴
If a cell grows on minimal medium, then it can synthesise
tryptophan.
Cell cannot grow on minimal medium.
Cell cannot synthesise tryptophan.
Given the rule P Q, and the fact P, infer the fact Q (abduction)
Deduction is sound in the logic we use (first-order predicate logic). Informally, this
means that if the rule and fact used in inference of the above form are true, then the
inferred fact must also be true. However, abduction is generally not sound. Thus, in the
abduction example, there could be many other reasons why the cell cannot grow.
Despite this, abduction is required to infer new scientific knowledge.
2.
Auxotrophic Growth Experiments and the Aromatic Amino Acid Pathway
The mutants (of strain BY4741 [ATCC201388] MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0;
Brachmann, C.B. et al. Designer deletion strains derived from Saccharomyces
cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disrution
and other applications. Yeast 14, 115-132) had the complete reading-frame of each
protein-encoding gene deleted by replacement with a selectable marker gene that has no
phenotype in the absence of the selective agent (Winzeler, E.A. et al. Functional
characterization of the S. cerevisiae genome by gene deletion and parallel analysis.
Science 285, 901-906, 1999; Giaever G. et al. Functional profiling of the
Saccharomyces cerevisiae genome Nature 418, 387-391, 2002). They are thus nonrevertible null mutants. Limitations in the availability of mutant yeast strains restricted
the number of genes used in the in vivo investigations to fifteen, and the auxotrophic
experimental requirement for a difference in growth phenotype, in turn, reduced this
number to eight (the other mutants always either grew or failed to grow): ybr166c,
ydr007w, ydr035w, ydr354w, yer090w, ygl026c, ykl211c, ynl316c. The number of
possible metabolites was limited by availability and cost to nine: anthranilate (10),
indole (190), p-hydroxyphenol pyruvic acid (193), L-phenylalanine (53),
phenylpyruvate (30), phosphoenol pyruvate (9385), shikimic acid (633), L-tyrosine
(53), L-tryptophan (53). The numbers in brackets are the normalised true experimental
costs of using each metabolite in a growth medium, note the ~3 order of magnitude
range.
Note that, in this pathway, some open reading-frames (ORFs) encode enzymes
that catalyse more than one biochemical reaction (e.g. YDR127w – 3-dehyrdoquinate
synthase, 3-dehydroquinate dehydratase, shikamate 5-dehydroenase, shikimate kinase,
3- phosphoshikimate 1-carboxyvinyltransferase); while, for other reactions, there are
iso-enzymes encoded by different ORFs (e.g. YBR249c and YDR035w both encode
phospho-2-dehydro-3-deoxyheptonate aldolase), thus providing redundancy.
Investigating the full behavior of the available genes and metabolites would
require at least 7,665 growth experiments (without repetition). We therefore decided to
restrict the investigation to experiments with either a single metabolite or a pair of
metabolites added. The number of experiments for each gene is thus restricted to 45 (9
+ ((9*8)/2)), giving 360 (8*45) possible experiments. A single experiment uses a
single well of the 96-well plate. One [mutant + medium] combination is placed in each
of the 8 wells, in a single column, together with minimal medium agar gel. The
mutants are pre-grown overnight in 5ml rich medium in a shaker at 37ºC (200rpm) then
diluted to a 1:100 concentration in a ¼ strength Ringer’s solution, whilst the added
metabolites are made up to a 0.2% concentration. As a control, each medium
combination found on a single plate is also used with the wild-type yeast (BY4741) that
is the parent of the mutant strains. To reduce (and to monitor) contamination, wells
belonging to the outer columns of the plate were filled with agar only. After the
experiments were set up, the plates were incubated for 24 hours at 30ºC and then
growth measured. Growth was measured using a Wallac 1420 Multilabel counter. The
mean and median growth measurement for each experimental combination (mutant +
medium) is derived from all wells containing the combination on the plate (usually 8
wells, but 16 if two columns have the same combination of substances). The following
simple decision tree was used to determine growth (see below for an explanation of
how this was formed):
If the mean of the mutant’s growth =< 0.44225 then the growth class is “no-growth” else
If the difference between the median values for the mutant’s growth and the wild type’s =< 0.2565 then the growth class is “no-growth” else the growth class is “growth”.
The use of this tree is a form of inductive inference.
3.
Computational Model of the Aromatic Amino Acid Pathway
The Prolog model of the pathway was refined in three stages:
The original model was translated from KEGG and carefully checked with the
literature. We checked that all the KEGG reactions were documented in S.
cerevisiae (consistency), and that there were no other related reactions described in
the literature (completeness).
The predictions of the model were then compared with the results of the singlemetabolite experiments (see above). Whether growth or no-growth was observed
was at this point decided visually. Since certain metabolites did not seem to affect
growth in the way predicted by the literature, we refined the model to make these
metabolites unable to be imported into the cells efficiently. This inference was, of
course, an abduction. It was also necessary to add inhibition effects. For example,
the results for adding tyrosine to ydr035w deletion mutants were anomalous: without
tyrosine, the mutants grew; with tyrosine they didn’t. This was unexpected, as one
would predict that the result of adding an amino acid, such as tyrosine, should be
monotonic as regards to growth. Our implemented explanation of this is that
YBR249C and YDR035W encode isoenzymes that catalyse the reaction:
phosphoenolpyruvate + erythrose 4-phosphoric acid -> 7-P-2-dehydro-3-deoxy-darabino-heptonate; when YDR035W is deleted, YBR249C remains and allows
pathways in the graph to tyrosine, tryptophan, and phenyalalanine. In the presence
of tyrosine in the medium, the enzymic product of YBR249C is inhibited, blocking
the pathway to tryptophan and phenyalalanine, and stopping growth.
The results of the double-metabolite experiments were then tested against the model,
and the automatic growth-calling software optimised by learning a decision tree to fit
the experimental results to the model (see above).The model developed on the single
metabolites was consistent with all but < 1.5% of the double-metabolite experiments.
The model was not further changed to include these experimental discrepancies. The
final model is therefore logically “incorrect”, in that it incorrectly predicts the results of
some experimental observations. We consider this a “feature” of the model, as it is the
typical situation in biological research.
There are two types of “noise” in the physical experiments:
Experiment and measurement noise (we estimate that ~25% of experiments are
noisy, i.e. they give an observation of growth or no-growth different from expected).
Noise due to errors in the background knowledge (the model does not agree with <
1.5% of experimental results.)
Because of the possibility of noise, we also implemented a simple system to allow the
Robot Scientist to backtrack when all possible hypotheses were contradicted by
experimental results. A training set is known to contain misclassified examples when
either of two situations occurs: the hypotheses generated at a given iteration are not a
subset of the hypotheses generated at the previous iteration, or no hypotheses can be
generated at a given iteration. In an effort to correct the misclassified training
examples, new training sets are generated where the classification of a single example
is changed. Hypotheses are generated from each of these new training sets and the
hypothesis set containing the fewest hypotheses is chosen. This noise abatement
system has a number of drawbacks: it assumes that there is only one misclassified
example in a training set and the process of altering the classification of training
examples does not guarantee that the new hypothesis set is correct.
4.
Performance Measures
The average performance of the hypotheses is an appropriate performance
measure because it rewards learners that discriminate between competing hypotheses.
This approach is a compromise between selecting the highest probability hypothesis,
and weighting all predictions by the probability of the hypotheses that generated them.
In active learning, the performance curves that have been generally used plot predictive
accuracy against the number of training examples. Often, two curves are plotted on the
same graph, one for active learning and one for random sampling. The accuracy of a
single hypothesis is the number of correct predictions that this hypothesis makes about
all possible single- and double-metabolite experiments, based on using the model as the
oracle. (An alternative approach would have been to have used the hand-generated
expected result of the experiments, but this has the disadvantage of not giving the
correct hypothesis 100% accuracy, as well as compromising the status of the Robot
Scientist as an automated system.) Such performance plots allow the difference in the
number of experiments (examples/time) required to reach a particular level of
performance to be compared. However, one drawback of such plots is that they ignore
any variation in the price of obtaining individual examples. When such variation does
exist, and the aim is to compare the price of attaining particular levels of performance,
these plots are potentially misleading. To overcome this drawback, we also plot the
cumulative price of the experiments against performance (Bryant et al., 2001). For this,
we use the normalised price of the metabolite. At the start of the experiments, when
there are 8 possible hypotheses, the average accuracy is 57%.
5.
Structure of the Robot Scientist
The robot automates the task of liquid handling and can conduct assays by
pipetting and mixing liquids on microtitre plates. The robot is controlled using TCL,
and we have written a compiler that translates Prolog commands into TCL robot
operations. Given a Prolog definition of one or more experiments, we have developed
code which designs a layout of the robot that will allow these experiments, with
controls, to be carried out efficiently. In addition, the robot has to be automatically
programmed to plate out the yeast and media into the correctly defined wells. The
microtitre plates were measured using the adjacent plate reader and the results were
returned to the LIMS. To reduce cost, the transfer of the plates from the robot to the
incubator, and from the incubator to the plate reader were done manually – although
this would have been trivial to automate. The key point is that there was no human
intellectual input in the experiment design/interpretation etc.
6.
Prolog Model
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% This model is designed to mimic auxotrophic mutant experiments in
% the aromatic amino acid pathway of yeast. It pertains to
% Phenylalanine, tyrosine and tryptophan biosynthesis. See KEGG map
% 400 at
% http://www.genome.ad.jp/dbget-bin/get_pathway?org_name=sce&mapno=00400
% Also see Stryer Chp 28 page 724.
% Note that the pathway had to be carefully checked by hand as there
% were errors and missing data in KEGG. The model is a
% representation of all of the known steps in this pathway.
% The model does not include genes as such. Instead it includes Open
% Reading Frames (ORFs) which are putative genes. Some ORFs may not
% code for anything.
% The code for processing sets assumes that any lists are ordered.
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
start('C00631').
start('C00279').
start('C00005').
start('C00000'). % h not in KEGG
start('C00002').
start('C00014').
start('C00064').
start('C00119').
start('C00065').
start('C00003').
start('C00006').
start('C00001'). % h2o
start('C00011'). % co2
start('C00025').
end('C00078').
end('C00079').
end('C00082').
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% Import metabolites
% To account for slow import of some metabolites
% NB use of "I" to label metabolites outside cell
enzyme(i1,[import],[x],1,1,[['I00074']],[['C00074']]).
enzyme(i2,[import],[x],1,1,[['I00078']],[['C00078']]).
enzyme(i3,[import],[x],1,1,[['I00079']],[['C00079']]).
enzyme(i4,[import],[x],1,1,[['I00082']],[['C00082']]).
enzyme(i5,[import],[x],1,1,[['I00108']],[['C00108']]).
enzyme(i6,[import],[x],1,2,[['I00166']],[['C00166']]). % slow
enzyme(i7,[import],[x],1,1,[['I00463']],[['C00463']]).
enzyme(i8,[import],[x],1,1,[['I00493']],[['C00493']]).
enzyme(i9,[import],[x],1,2,[['I01179']],[['C01179']]). % slow
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% Enzymes in aromatic pathway
% enzyme(+enz_id,[+orf|ORFS],[+enz_class|Classes],+direction,+day,
%
[+lhs|Lefts],[+rhs|Rights]).
% enz_id - unique ID referencing enzyme classes
% orf - open reading frame
% enz_class - a list of enzyme classes. Each class is the Enzyme
%
Commission classification number for an enzyme.
% lhs - [+left|LHS] - a list of metabolites to be found in the lhs of
% a reaction
%
% rhs - [+right|RHS] a list of metabolites to be found in the rhs of
% a reaction
enzyme(e1,['YGR254W'],['4.2.1.11'],1,1,[['C00631']],[['C00001','C00074']]).
enzyme(e2,['YHR174W'],['4.2.1.11'],1,1,[['C00631']],[['C00001','C00074']]).
enzyme(e3,['YMR323W'],['4.2.1.11'],1,1,[['C00631']],[['C00001','C00074']]).
enzyme(e4,['YBR249C'],['4.1.2.15'],1,1,[['C00001','C00074','C00279']],
[['C00009','C04691']]).
enzyme(e5,['YDR035W'],['4.1.2.15'],1,1,[['C00001','C00074','C00279']],
[['C00009','C04691']]).
enzyme(e6,['YDR127W'],['4.6.1.3','4.2.1.10','X','1.1.1.25','2.7.1.71','2.5.1.19'],
1,1,[['C04691'],['C00944'],['C02637'],['C00000','C00005','C02652'],
['C00002','C00493'],['C00074','C03175']],
[['C00009','C00944'],['C00001','C02637'],['C02652'],['C00006','C00493'],
['C00008','C03175'],['C00009','C01269']]).
enzyme(e7,['YGL148W'],['4.6.1.4'],1,1,[['C01269']],[['C00009','C00251']]).
enzyme(e8,['YER090W'],['4.1.3.27'],1,1,[['C00014','C00251']],
[['C00001','C00022','C00108']]).
enzyme(e9,['YER090W','YKL211C'],['4.1.3.27'],1,1,[['C00064','C00251']],
[['C00022','C00025','C00108']]).
enzyme(e10,['YDR354W'],['2.4.2.18'],1,1,[['C00108','C00119']],
[['C00013','C04302']]).
enzyme(e11,['YDR007W'],['5.3.1.24'],1,1,[['C04302']],[['C01302']]).
enzyme(e12,['YKL211C'],['4.1.1.48'],1,1,[['C01302']],
[['C00001','C00011','C03506']]).
enzyme(e13,['YGL026C'],['4.2.1.20'],1,1,[['C00065','C03506'],['C03506'],
['C00065','C00463']],[['C00001','C00078','C00661'],
['C00463','C00661'],['C00001','C00078']]).
enzyme(e14,['YPR060C'],['5.4.99.5'],1,1,[['C00251']],[['C00254']]).
enzyme(e15,['YNL316C'],['4.2.1.51'],1,1,[['C00003','C00254']],
[['C00004','C00011','C00166']]).
enzyme(e16,['YBR166C'],['1.3.1.13'],1,1,[['C00006','C00254']],
[['C00000','C00005','C00011','C01179']]).
enzyme(e17,['YGL202W'],['2.6.1.7'],1,1,[['C00025','C00166'],['C00025','C01179']],
[['C00026','C00079'],['C00026','C00082']]). %not in KEGG
enzyme(e18,['YHR137W'],['2.6.1.7'],1,1,[['C00025','C00166'],['C00025','C01179']],
[['C00026','C00079'],['C00026','C00082']]). %not in KEGG
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% inhibitor(+orf,[+enz_id|Enzymes],+enz_class,[+met|Metabolites])
% inhibitor/4 states that each of the given list of metabolites
% inhibits the function of the enzyme(s) coded for by the given ORF
% orf - open reading fram
% enz_id - unique enzyme id number
% enz_class - Enzyme Commision enzyme class
% met - metabolite number
inhibitor('YBR249C',[e4],'4.1.2.15',['C00082']).
inhibitor('YDR035W',[],'4.1.2.15',[]).
inhibitor('YDR127W',[],'1.1.1.25',[]).
inhibitor('YGL148W',[],'4.6.1.4',[]).
inhibitor('YER090W',[],'4.1.3.27',[]).
inhibitor('YKL211C',[],'4.1.3.27',[]).
inhibitor('YGR354W',[],'4.2.1.11',[]).
inhibitor('YDR007W',[],'5.3.1.24',[]).
inhibitor('YGL026C',[],'4.2.1.20',[]).
inhibitor('YPR060C',[],'5.4.99.5',[]).
inhibitor('YNL316C',[],'4.2.1.51',[]).
inhibitor('YGL202W',[],'2.6.1.7',[]).
inhibitor('YHR137W',[],'2.6.1.7',[]).
inhibitor('YBR166C',[],'1.3.1.13',[]).
% shared_orfs(+orf,[+enz_id|Enzymes])
% The given ORF is involved in the production of the given list of
% enzymes (must be more than one - the enzymes ``share'' the ORF)
shared_orfs('YER090W',[e8,e9]).
shared_orfs('YKL211C',[e9,e12]).
% new_enzyme/4 - test the reaction coded for by the enzyme
% the enzyme should not be used as an import mechanism
new_enzyme(EnzId,EnzClass,Reactants,Products) :enzyme(EnzId,Orfs,EnzClass,_Direction,_Day,Reactants,Products),
\+Orfs == [import].
% change_all_metabolites changes all import metabolite identifying
% labels from C**** to I***** e.g C00074 becomes I00074
% used to convert metabolites appearing in 2nd argument of phenotypic_effect/2
change_all_metabolites([],RDone,Changed) :reverse(RDone,Changed).
change_all_metabolites([M|Metabolites],Done,Changed) :change_metabolite(M,CM),!,
change_all_metabolites(Metabolites,[CM|Done],Changed).
change_metabolite(Metabolite,NewMetabolite) :name(Metabolite,[_C|Ascii]),
conc([73],Ascii,NewAscii),
name(NewMetabolite,NewAscii).
% inhibited/2 - test whether an enzyme is inhibited by the
% current cell contents
inhibited(EnzId1,Starts) :inhibitor(_Orf,EnzIds,_,List),
member(EnzId1,EnzIds),
member(Inhib,Starts),
member(Inhib,List).
same_enzyme(EnzId1,EnzId2) :EnzId1 == EnzId2.
% same_enzyme_function/9 is true if those metabolites on the RHS
% that belong to the main pathway are the same for each enzyme,
% A metabolite belongs to the main pathway if it appears in the
% starts, is an end product, or appears in the LHS lists of the enzyme
% definitions.
% assumes only forwards direction - could be made reversible if
% direction was taken into account and the RHS was used when the
% system ran backwards
same_enzyme_function(EnzClass1,EnzClass2,
Lefts,_LHS1,RHS1,_LHS2,RHS2) :EnzClass1 == EnzClass2,
main_pathway_rights(RHS1,Lefts,[],Mains1),
main_pathway_rights(RHS2,Lefts,[],Mains2),
quicksort_mets(Mains1,SortedMains1),
quicksort_mets(Mains2,SortedMains2),
SortedMains1 == SortedMains2.
%write(Enzyme:Enzyme2),nl,
%write(Reactants:R1),nl,
%write(Products:R2),nl,%get0(10).
%write('evaluate_enzyme succeeded'),nl.
% gene_sharing/2 tests to see if an enzyme (enz_id) can be made by two
% or more genes (shared)
gene_sharing(EnzId1,EnzId2) :shared_orfs(_Gene,EnzIds),
member(EnzId1,EnzIds),
member(EnzId2,EnzIds).
% An enzyme can't be used if is the same enzyme as those produced
% by the knocked out orf (including those enzymes ``shared'' - i.e.
% where two or more orfs are required to produce the enzyme)
cant_use_enzyme(EnzId1,EnzId2,EnzClass1,EnzClass2,
Lefts,_LHS1,RHS1,_LHS2,RHS2) :same_enzyme_function(EnzClass1,EnzClass2,
Lefts,_LHS1,RHS1,_LHS2,RHS2),
same_enzyme(EnzId1,EnzId2).
cant_use_enzyme(EnzId1,EnzId2,EnzClass1,EnzClass2,
Lefts,_LHS1,RHS1,_LHS2,RHS2) :same_enzyme_function(EnzClass1,EnzClass2,
Lefts,_LHS1,RHS1,_LHS2,RHS2),
gene_sharing(EnzId1,EnzId2).
% uses Transitive Closure
% All Metabolites added to the minimal media are converted to import
% metabolites and added to the starts. Pathway reactions are
% tested by evaluating each enzyme at a time - deciding whether
% the enzyme is available (i.e not produced by a knocked out ORF)
% and adding any metabolites formed by the reaction.
% backtracking ensures all (allowable) reactions are tested. If
% the end products are not found in the results from this process
% the procedure succeeds i.e. phenotypic_effect/2 succeeds if
% there is NO GROWTH for the given ORF and nutrients (metabolites
% added to the minimal media)
% When the model is used in hypotheses generation, the encodes/5
% fact is constructed by the learning step and appears in the
% hypotheses generated (the codes/7 facts are omitted from the model)
phenotypic_effect(ORF,Nutrients):change_all_metabolites(Nutrients,[],Changed),
bagof(S,start(S),Minimal),
union(Minimal,Changed,Starts),
%write('Starts ':Starts),nl,
bagof(End,end(End),Ends),
find_knocked_out_enzymes(ORF,KnockedOut),
all_lefts(Lefts),
%write(KnockedOut),get0(10),nl,
new_enzyme(EnzId,EnzClass,LHS,RHS),
%write(EnzId:EnzClass:LHS:RHS),nl,
encodes(ORF,EnzId,EnzClass,LHS,RHS),
%write(ORF:EnzClass:LHS:RHS),nl,!,
connected_without_this_step(Starts,MutantProducts,
EnzId,EnzClass,Lefts,LHS,RHS,1),
%write('Mutant Products ':MutantProducts),nl,
\+new_subset(Ends,MutantProducts).
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% connected_without_this_step/5 succeeds if all of the end points can
% be reached from the start points without involving the reaction
% specified in the 3rd,4th and 5th terms.
connected_without_this_step(Starts1,Ends,EnzId,EnzClass,
Lefts,Reactants,Products,Day) :expand_without_this_step(Starts1,Starts2,EnzId,EnzClass,
Lefts,Reactants,Products,Day),!,
connected_without_this_step(Starts2,Ends,EnzId,EnzClass,
Lefts,Reactants,Products,Day).
connected_without_this_step(Starts,Starts,_EnzId,_EnzClass,
_Lefts,_Reactants,_Products,_Day).
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% expand_without_this_step/9 attempts to add new metabolites from a
% reaction, checking that the enzyme catalysing the reaction is ...
%
%
1) Not inhibited by any of the current cell products
2) Available for use (not coded for by the knocked out gene)
expand_without_this_step(Starts1,Starts2,EnzId1,EnzClass1,Lefts,
Reactants,Products,Day) :enzyme1(EnzId2,_Orfs,EnzClass2,R1,R2,Starts1,Day),
%write(EnzId2:EnzClass2:R1:R2),nl,
%write(KnockedOut),nl,
\+inhibited(EnzId2,Starts1),
\+cant_use_enzyme(EnzId1,EnzId2,EnzClass1,EnzClass2,
Lefts,Reactants,Products,R1,R2),
valid_reactants(R1,R2,Starts1,1,RHS),
union(RHS,Starts1,Starts2).
%write('Adding ':RHS),get0(10),nl.
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% subset(Set, Subset).
subset([], []):!.
subset([H | T], [H | T2]):!,
subset(T, T2).
subset([H | T], T2):subset(T, T2).
new_subset([],_S).
new_subset([I|Items],Set) :member(I,Set),
new_subset(Items,Set).
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%
% set_union/3 - set union for ordered lists.
%
union([],S2,S2) :- !.
union(S1,[],S1) :- !.
union([H1|T1],[H2|T2],[H1|T3]) :H1 == H2,
union(T1,T2,T3), !.
% Equal
union([H1|T1],[H2|T2],[H2|T3]) :H1 @> H2, !,
union([H1|T1],T2,T3), !.
% Greater than
union([H1|T1],S2,[H1|T3]) :- !,
union(T1,S2,T3), !.
% Less than
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
ends(['C00078', 'C00079', 'C00082']).
% On minimal medium need to have a path from starts to all ends
% because every end point is needed for growth.
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
generated_by_other_pathways(['C00000', 'C00001', 'C00002', 'C00003',
'C00005', 'C00006', 'C00014', 'C00025', 'C00064', 'C00065',
'C00119', 'C00279', 'C00631']).
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% mandatory_nutrients/1
mandatory_nutrients([]).
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% optional_nutrients/1
optional_nutrients(['C00074', 'C00078', 'C00079', 'C00082', 'C00108',
'C00166', 'C00463', 'C00493', 'C01179']).
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% Ignoring Direction and Day:
% Direction is always forward only
% Only condiering Day = 1
encodes(ORF,EnzId,Type,Reactants1,Reactants2):codes(ORF,EnzId,Type,Direction,Day,Reactants1,Reactants2).
%enzyme(EnzId,Orfs,Type, Reactants1, Reactants2):%
enzyme(EnzId,Orfs,Type, Direction, Day, Reactants1, Reactants2).
enzyme1(EnzId,Orfs,Enzyme,R1,R2,Starts,Day) :enz(EnzId,Orfs,Enzyme,R1,R2,Day).
enz(EnzId,Orfs,Ec,R1,R2,Day) :enzyme(EnzId,Orfs,Ec,1,D,R1,R2),
D =< Day.
% tests time
% non reverse
enz(EnzId,Orfs,Ec,R1,R2,Day) :enzyme(EnzId,Orfs,Ec,2,D,R1,R2),
D =< Day.
% reverse
enz(EnzId,Orfs,Ec,R2,R1,Day) :enzyme(EnzId,Orfs,Ec,2,D,R1,R2),
D =< Day.
% reverse
% valid_reactants/5 ensures that a new reaction is evaluated when an
% enzyme evaluated - used when more than one reaction is coded for by
% one ORF e.g. YDR127W
% i.e. all new metabolites are are eventually added
valid_reactants([LHS|_Lefts],Rights,Starts,I,RHS) :new_subset(LHS,Starts),
find_rhs(1,I,Rights,RHS),
\+new_subset(RHS,Starts).
valid_reactants([_LHS|Lefts],Rights,Starts,I,RHS) :I2 is I + 1,
valid_reactants(Lefts,Rights,Starts,I2,RHS).
find_rhs(I,N,[RHS|_Rights],RHS) :I == N.
find_rhs(I,N,[_RHS|Rights],Found) :I2 is I + 1,
find_rhs(I2,N,Rights,Found).
in_mets_list(Metabolite,[MList|_LHS]) :member(Metabolite,MList).
in_mets_list(Metabolite,[_MList|LHS]) :in_mets_list(Metabolite,LHS).
in_lhs(Metabolite,[LHS|_Lefts]) :in_mets_list(Metabolite,LHS).
in_lhs(Metabolite,[_LHS|Lefts]) :in_lhs(Metabolite,Lefts).
main_pathway(Metabolite,_Lefts) :start(Metabolite).
main_pathway(Metabolite,Lefts) :in_lhs(Metabolite,Lefts).
main_pathway(Metabolite,_Lefts) :end(Metabolite).
all_lefts(Lefts) :bagof(LHS,EId^Genes^EC^Dir^Day^RHS^enzyme(EId,Genes,EC,Dir,Day,LHS,RHS),Lefts).
main_pathway_rhs([],_Lefts,Mains,Mains).
main_pathway_rhs([Met|Metabolites],Lefts,Done,Mains) :main_pathway(Met,Lefts),
main_pathway_rhs(Metabolites,Lefts,[Met|Done],Mains).
main_pathway_rhs([_Met|Metabolites],Lefts,Done,Mains) :main_pathway_rhs(Metabolites,Lefts,Done,Mains).
main_pathway_rights([],_Lefts,Mains,Mains).
main_pathway_rights([RHS|Rights],Lefts,Done,Mains) :main_pathway_rhs(RHS,Lefts,[],Found),
conc(Done,Found,NewDone),
main_pathway_rights(Rights,Lefts,NewDone,Mains).
member(I,[I|_Items]).
member(I,[_OI|Items]) :member(I,Items).
conc([],L,L).
conc([X|L1],L2,[X|L3]) :conc(L1,L2,L3).
reverse([],[]).
reverse([I],[I]).
reverse([I1,I2],[I2,I1]).
reverse([I|Items],ReversedRest) :reverse(Items,Reversed),
conc(Reversed,[I],ReversedRest).
quicksort_mets([],[]).
quicksort_mets([X|Tail],Sorted) :split_mets(X,Tail,Small,Big),
quicksort_mets(Small,SortedSmall),
quicksort_mets(Big,SortedBig),
append(SortedSmall,[X|SortedBig],Sorted).
split_mets(_,[],[],[]).
split_mets(X,[Y|Tail],[Y|Small],Big) :X @> Y,!,
split_mets(X,Tail,Small,Big).
split_mets(X,[Y|Tail],Small,[Y|Big]) :split_mets(X,Tail,Small,Big).
find_all_enzymes(Enzymes) :bagof(enzyme(EnzId,Orfs,EnzClass,Direction,Day,LHS,RHS),
enzyme(EnzId,Orfs,EnzClass,Direction,Day,LHS,RHS),Enzymes).
knocked_out_enzymes(_Orf,[],KnockedOut,KnockedOut).
knocked_out_enzymes(Orf,[Enz|Enzymes],Done,KnockedOut) :Enz = enzyme(EnzId,Orfs,_EnzClass,_Direction,_Day,_LHS,_RHS),
member(Orf,Orfs),
knocked_out_enzymes(Orf,Enzymes,[EnzId|Done],KnockedOut).
knocked_out_enzymes(Orf,[_Enz|Enzymes],Done,KnockedOut) :knocked_out_enzymes(Orf,Enzymes,Done,KnockedOut).
find_knocked_out_enzymes(Orf,KnockedOut) :find_all_enzymes(Enzymes),
knocked_out_enzymes(Orf,Enzymes,[],KnockedOut).
knocked_out_enzyme(EnzId,KnockedOut) :member(EnzId,KnockedOut).
7.
Experimental Data
Iteration ase
0
1
2
3
4
5
random
57.36
67.17
76.14
79.54
80.47
80.11
naive
57.36
67.39
72.59
71.58
73.03
72.16
57.36
67.27
68.55
73.83
73.89
73.94
Table 1: Classification Accuracies for ase, random and naïve. Experimental Data
Iteration ase
0
1
2
3
4
5
random
0.00
1.00
1.76
2.26
2.41
2.50
naive
0.00
2.91
3.56
3.75
3.87
3.96
0.00
1.00
1.59
1.89
2.11
2.26
Table 2: Relative Cost (Log10 £) for ase, random and naïve. Experimental Data
Ase
Gene
Run
Technique
Iteration
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
Cost
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
Accuracy
69.3827
74.3210
28.8889
74.3210
59.7531
79.2593
63.7427
69.3827
69.3827
63.7427
53.4503
74.3210
59.7531
79.2593
74.3210
69.3827
69.3827
74.3210
53.4503
74.3210
59.7531
79.2593
74.3210
69.3827
69.3827
74.3210
53.4503
74.3210
59.7531
79.2593
74.3210
47.8363
Table 3: Ase Results for the Robot Scientist: Iteration 1. Experimental Data
Ase
Gene
Run
Technique
Iteration
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
Cost
62
62
62
62
62
62
39
62
62
39
39
62
62
62
62
62
62
62
39
62
62
62
62
62
62
62
39
62
62
62
62
39
Accuracy
69.3827
95.5556
43.3333
95.5556
80.0000
86.6667
88.3333
69.3827
69.3827
88.3333
53.4503
95.5556
80.0000
86.6667
95.5556
69.3827
69.3827
95.5556
55.0000
95.5556
59.7531
79.2593
74.3210
69.3827
69.3827
95.5556
55.0000
95.5556
59.7531
79.2593
74.3210
42.7778
Table 4: Ase Results for the Robot Scientist: Iteration 2. Experimental Data
Ase
Gene
Run
Technique
Iteration
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
Cost
166
251
251
251
251
251
78
166
166
78
78
251
251
251
251
166
166
251
78
251
166
166
166
166
166
251
78
251
166
166
166
78
Accuracy
69.3827
100.0000
46.6667
100.0000
84.4444
86.6667
88.3333
82.2222
82.2222
88.3333
55.0000
100.0000
80.0000
86.6667
100.0000
82.2222
82.2222
100.0000
55.0000
100.0000
59.7531
79.2593
74.3210
69.3827
82.2222
100.0000
55.0000
100.0000
59.7531
79.2593
74.3210
42.7778
Table 5: Ase Results for the Robot Scientist: Iteration 3. Experimental Data
Ase
Gene
Run
Technique
Iteration
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
Cost
270
280
280
280
280
450
130
218
218
130
130
280
450
450
280
218
218
280
130
280
270
270
270
270
218
280
130
280
270
270
270
130
Accuracy
63.5556
100.0000
46.6667
100.0000
84.4444
86.6667
88.3333
82.2222
100.0000
88.3333
55.0000
100.0000
80.0000
86.6667
100.0000
82.2222
82.2222
100.0000
55.0000
100.0000
59.7531
79.2593
74.3210
69.3827
100.0000
100.0000
55.0000
100.0000
59.7531
79.2593
74.3210
42.7778
Table 6: Ase Results for the Robot Scientist: Iteration 4. Experimental Data
Ase
Gene
Run
Technique
Iteration
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
ase
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
Cost
459
319
319
319
319
479
182
270
218
182
182
319
479
479
319
270
270
319
182
319
374
374
374
374
218
319
182
319
374
374
374
182
Accuracy
63.5556
100.0000
46.6667
100.0000
84.4444
86.6667
88.3333
82.2222
100.0000
88.3333
55.0000
88.3333
80.0000
86.6667
100.0000
82.2222
82.2222
100.0000
55.0000
100.0000
59.7531
79.2593
74.3210
69.3827
100.0000
100.0000
55.0000
100.0000
59.7531
79.2593
74.3210
42.7778
Table 7: Ase Results for the Robot Scientist: Iteration 5. Experimental Data
Random
Gene
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
Run
Technique
Iteration
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
Cost
247
81
62
247
104
62
241
205
189
822
62
9427
9427
241
104
224
81
29
62
62
104
685
62
384
224
104
199
643
241
247
218
685
Accuracy
81.3333
79.4444
53.4503
76.4646
63.1579
81.1111
63.7427
69.3827
78.5185
63.7427
53.4503
76.4646
67.9798
80.4444
63.7427
64.5455
64.5556
76.8687
53.4503
77.2222
47.1111
56.9591
77.2222
78.5185
64.5455
81.1111
20.0000
74.3210
63.1579
74.8485
63.7427
64.5556
Table 8: Random Results for the Robot Scientist: Iteration 1. Experimental Data
Random
Gene
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
.YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
Run Technique
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
Iteration
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
Cost
488
133
257
446
309
9626
9626
404
388
10249
143
18831
9531
293
9531
276
276
276
143
724
9489
880
309
9788
305
299
9603
861
9668
328
1046
926
Accuracy
81.3333
93.1481
53.4503
76.4646
72.0000
81.1111
95.5556
69.3827
78.5185
63.7427
53.4503
76.4646
85.3704
80.4444
82.0833
64.5455
64.5556
76.8687
17.3333
77.2222
47.1111
81.2963
77.2222
78.5185
100.0000
81.1111
20.0000
74.3210
85.3704
74.8485
95.3333
64.5556
Table 9: Random Results for the Robot Scientist: Iteration 2. Experimental Data
Random
Gene
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
Run Technique
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
Iteration
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
Cost
687
374
481
670
498
9831
10311
9831
1050
10353
828
19474
9593
9678
18916
328
523
328
9570
786
10132
942
319
19215
344
984
10288
1079
10353
546
10610
1611
Accuracy
81.3333
93.1481
53.4503
46.6667
84.4444
81.1111
95.5556
69.3827
78.5185
87.2222
47.2222
74.4444
85.3704
80.4444
74.4444
64.5455
64.5556
51.1111
17.3333
75.9259
47.1111
81.2963
77.2222
78.5185
100.0000
46.6667
33.3333
100.0000
85.3704
74.8485
95.3333
64.5556
Table 10: Random Results for the Robot Scientist: Iteration 3. Experimental Data
Random
Gene
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
Run Technique
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
Iteration
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
Cost
10251
1059
728
670
1160
9860
10363
10474
1735
11038
10255
19715
9622
10062
19601
990
9950
328
9769
838
10373
1627
371
19456
344
984
10288
1463
19917
1179
10714
11038
Accuracy
81.3333
93.1481
100.0000
46.6667
84.4444
81.1111
95.5556
69.3827
78.5185
87.2222
47.2222
74.4444
85.3704
80.4444
74.4444
64.5455
64.5556
51.1111
17.3333
75.9259
47.1111
81.2963
77.2222
78.5185
100.0000
46.6667
33.3333
100.0000
85.3704
74.8485
95.3333
64.5556
Table 11: Random Results for the Robot Scientist: Iteration 4. Experimental Data
Random
Gene
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
Run Technique
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
random
Iteration
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
Cost
10303
10623
728
670
10545
10049
10425
11136
2378
11723
10359
19796
9869
10114
29005
1675
10054
328
9831
1037
10425
11012
560
19485
344
984
10288
1710
20579
10606
10776
11285
Accuracy
81.3333
93.1481
100.0000
46.6667
84.4444
81.1111
95.5556
69.3827
78.5185
87.2222
47.2222
46.6667
85.3704
80.4444
74.4444
64.5455
64.5556
51.1111
17.3333
75.9259
47.1111
81.2963
77.2222
78.5185
100.0000
46.6667
33.3333
100.0000
85.3704
74.8485
95.3333
64.5556
Table 12: Random Results for the Robot Scientist: Iteration 5. Experimental Data
Naive
Gene
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
Run
Technique
Iteration
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
Cost
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
Accuracy
69.3827
74.3210
28.8889
74.3210
59.7531
79.2593
63.7427
69.3827
69.3827
63.7427
53.4503
74.3210
59.7531
79.2593
74.3210
69.3827
69.3827
74.3210
53.4503
74.3210
59.7531
79.2593
74.3210
69.3827
69.3827
74.3210
56.4503
74.3210
59.7531
79.2593
74.3210
47.8363
Table 13: Naive Results for the Robot Scientist: Iteration 1. Experimental Data
Naive
Gene
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
Run
Technique
Iteration
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
Cost
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
39
Accuracy
69.3827
74.3210
28.8889
74.3210
59.7531
79.2593
88.3333
69.3827
69.3827
88.3333
53.4503
74.3210
59.7531
79.2593
74.3210
69.3827
69.3827
74.3210
53.4503
74.3210
59.7531
79.2593
74.3210
69.3827
69.3827
74.3210
53.4503
74.3210
59.7531
79.2593
74.3210
42.7778
Table 14: Naive Results for the Robot Scientist: Iteration 2. Experimental Data
Naive
Gene
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
Run
Technique
Iteration
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
3
Cost
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
78
Accuracy
69.3827
88.3333
55.0000
74.3210
96.1111
79.2593
88.3333
69.3827
69.3827
88.3333
53.4503
88.3333
96.1111
79.2593
88.3333
69.3827
69.3827
88.3333
53.4503
74.3210
59.7531
79.2593
74.3210
69.3827
69.3827
74.3210
53.4503
88.3333
59.7531
79.2593
74.3210
42.7778
Table 15: Naive Results for the Robot Scientist: Iteration 3. Experimental Data
Naive
Gene
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
Run
Technique
Iteration
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
4
Cost
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
130
Accuracy
69.3827
88.3333
55.0000
95.5556
96.1111
86.6667
88.3333
69.3827
69.3827
88.3333
53.4503
88.3333
96.1111
79.2593
88.3333
54.4444
69.3827
88.3333
55.0000
74.3210
59.7531
79.2593
74.3210
69.3827
54.4444
74.3210
55.0000
88.3333
59.7531
79.2593
74.3210
42.7778
Table 16: Naive Results for the Robot Scientist: Iteration 4. Experimental Data
Naive
Gene
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
YBR166C
YDR007W
YDR035W
YDR354W
YER090W
YGL026C
YKL211C
YNL316C
Run
Technique
Iteration
1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
naive
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
Table 17: Naive Results for the Robot Scientist: Iteration 5.
Cost
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
182
Accuracy
69.3827
88.3333
55.0000
95.5556
96.1111
86.6667
88.3333
69.3827
69.3827
88.3333
55.0000
88.3333
96.1111
79.2593
88.3333
54.4444
69.3827
88.3333
55.0000
74.3210
59.7531
79.2593
74.3210
69.3827
54.4444
74.3210
55.0000
88.3333
59.7531
79.2593
74.3210
42.7778
7
Simulation Data
itera
tech
median 2.5%
tion
nique
Accuracy
97.5%
(median (97.5%
- 2.5%)
median 2.5%
Cost
97.5%
-median)
(median (97.5%
- 2.5%)
- median)
0 ase
1 ase
2 ase
3 ase
4 ase
5 ase
6 ase
7 ase
8 ase
9 ase
10 ase
57.36
69.70
78.59
89.28
95.63
97.85
97.85
97.85
97.85
97.85
97.85
57.36
69.70
78.59
89.28
95.63
97.85
97.85
97.85
97.85
97.85
97.85
57.36
69.70
78.59
89.28
95.63
97.85
97.85
97.85
97.85
97.85
97.85
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
1.00
1.77
2.20
2.26
2.27
2.27
2.27
2.27
2.27
2.27
0.00
1.00
1.77
2.20
2.26
2.27
2.27
2.27
2.27
2.27
2.27
0.00
1.00
1.77
2.20
2.26
2.27
2.27
2.27
2.27
2.27
2.27
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0 random
1 random
2 random
3 random
4 random
5 random
6 random
7 random
8 random
9 random
10 random
57.36
67.29
75.38
80.81
84.90
86.77
87.82
89.53
90.97
91.57
92.89
57.36
62.04
66.65
70.24
72.23
73.03
75.31
75.31
79.58
79.58
79.58
57.36
72.67
87.08
89.41
92.89
95.58
96.01
96.01
96.59
96.99
96.99
0.00
5.24
8.73
10.56
12.67
13.73
12.51
14.21
11.38
11.99
13.31
0.00
5.38
11.70
8.61
7.99
8.81
8.19
6.48
5.63
5.42
4.10
0.00
2.67
3.16
3.42
3.57
3.69
3.79
3.87
3.92
3.96
4.00
0.00
2.14
2.64
2.98
3.12
3.17
3.19
3.42
3.46
3.46
3.49
0.00
3.39
3.69
3.94
4.05
4.14
4.18
4.22
4.27
4.29
4.32
0.00
0.53
0.52
0.43
0.45
0.52
0.60
0.45
0.46
0.50
0.51
0.00
0.72
0.53
0.52
0.48
0.44
0.38
0.34
0.35
0.33
0.32
0 naive
1 naive
2 naive
3 naive
4 naive
5 naive
6 naive
7 naive
8 naive
9 naive
10 naive
57.36
69.70
73.82
73.82
82.71
86.53
96.18
96.18
96.18
96.18
96.18
57.36
69.70
73.82
73.82
82.71
86.53
96.18
96.18
96.18
96.18
96.18
57.36
69.70
73.82
73.82
82.71
86.53
96.18
96.18
96.18
96.18
96.18
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
1.00
1.60
1.87
2.06
2.17
2.24
2.27
2.30
2.33
2.36
0.00
1.00
1.60
1.87
2.06
2.17
2.24
2.27
2.30
2.33
2.36
0.00
1.00
1.60
1.87
2.06
2.17
2.24
2.27
2.30
2.33
2.36
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
0.00
Table 18. Median, 2.5,97.5 percentiles for Classification Accuracy and Relative Experimental Cost
(Log10 £): 100 runs. Robot Scientist simulation with 0% noise.
itera tech
tion
Accuracy
median 2.5% 97.5%
(median (97.5%
nique
- 2.5%)
median
2.5%
Cost
97.5%
-median)
(median (97.5%
- 2.5%)
- median)
0 ase
1 ase
2 ase
3 ase
4 ase
5 ase
6 ase
7 ase
8 ase
9 ase
10 ase
57.36
65.68
72.35
77.49
80.22
82.17
83.28
84.72
84.72
85.07
84.19
57.36
58.08
63.79
61.81
66.57
66.29
67.99
66.60
66.29
70.88
70.63
57.36
69.70
78.04
87.92
91.74
92.71
94.02
96.39
96.39
97.85
97.85
0.00
7.61
8.56
15.69
13.65
15.88
15.29
18.13
18.43
14.19
13.56
0.00
4.02
5.69
10.42
11.52
10.54
10.74
11.67
11.67
12.78
13.66
0.00
1.00
1.76
2.17
2.34
2.40
2.43
2.46
2.49
2.51
2.51
0.00
1.00
1.72
2.07
2.24
2.28
2.30
2.32
2.34
2.35
2.36
0.00
1.00
1.79
2.29
2.45
2.53
2.58
2.63
2.67
2.70
2.71
0.00
0.00
0.04
0.10
0.10
0.12
0.13
0.14
0.15
0.16
0.16
0.00
0.00
0.03
0.12
0.11
0.13
0.14
0.16
0.18
0.19
0.20
0 random
1 random
2 random
3 random
4 random
5 random
6 random
7 random
8 random
9 random
10 random
57.36
64.15
68.45
70.10
72.18
71.80
71.09
69.35
69.54
69.51
69.24
57.36
55.4
56.62
55.99
55.99
53.76
51.39
50.57
50.57
50.57
50.57
57.36
72.52
80.96
84.93
88.61
89.63
88.65
89.90
89.90
92.10
92.10
0.00
8.75
11.83
14.11
16.19
18.04
19.68
18.78
18.97
18.94
18.67
0.00
8.37
12.51
14.83
16.43
17.84
17.59
20.55
20.37
22.58
22.86
0.00
2.56
3.11
3.43
3.63
3.77
3.85
3.91
3.98
4.03
4.08
0.00
1.91
2.65
2.94
3.09
3.21
3.31
3.35
3.39
3.40
3.41
0.00
3.00
3.57
3.89
4.00
4.13
4.17
4.24
4.28
4.31
4.34
0.00
0.65
0.46
0.48
0.54
0.57
0.54
0.57
0.59
0.63
0.67
0.00
0.43
0.46
0.46
0.37
0.36
0.33
0.32
0.30
0.28
0.27
0 naive
1 naive
2 naive
3 naive
4 naive
5 naive
6 naive
7 naive
8 naive
9 naive
10 naive
57.36
65.48
69.46
71.69
75.73
77.58
79.58
79.65
79.65
79.58
79.58
57.36
58.25
59.98
63.05
67.83
63.60
60.76
60.76
60.76
59.81
59.81
57.36
69.70
75.76
77.99
81.80
85.21
90.55
91.51
91.51
91.51
94.38
0.00
7.23
9.48
8.64
7.89
13.97
18.83
18.90
18.90
19.77
19.77
0.00
4.22
6.31
6.30
6.08
7.63
10.96
11.86
11.86
11.93
14.79
0.00
1.00
1.60
1.87
2.06
2.19
2.27
2.34
2.40
2.44
2.49
0.00
1.00
1.60
1.87
2.06
2.17
2.24
2.29
2.32
2.34
2.37
0.00
1.00
1.60
1.90
2.12
2.27
2.36
2.44
2.51
2.57
2.63
0.00
0.00
0.00
0.00
0.00
0.02
0.04
0.06
0.08
0.10
0.11
0.00
0.00
0.00
0.04
0.07
0.08
0.09
0.10
0.11
0.13
0.14
Table 19. Median, 2.5,97.5 percentiles for Classification Accuracy and Relative Experimental Cost
(Log10 £): 100 runs. Robot Scientist simulation with 25% noise: noise abatement strategy used