SP1 Protein production order form
advertisement
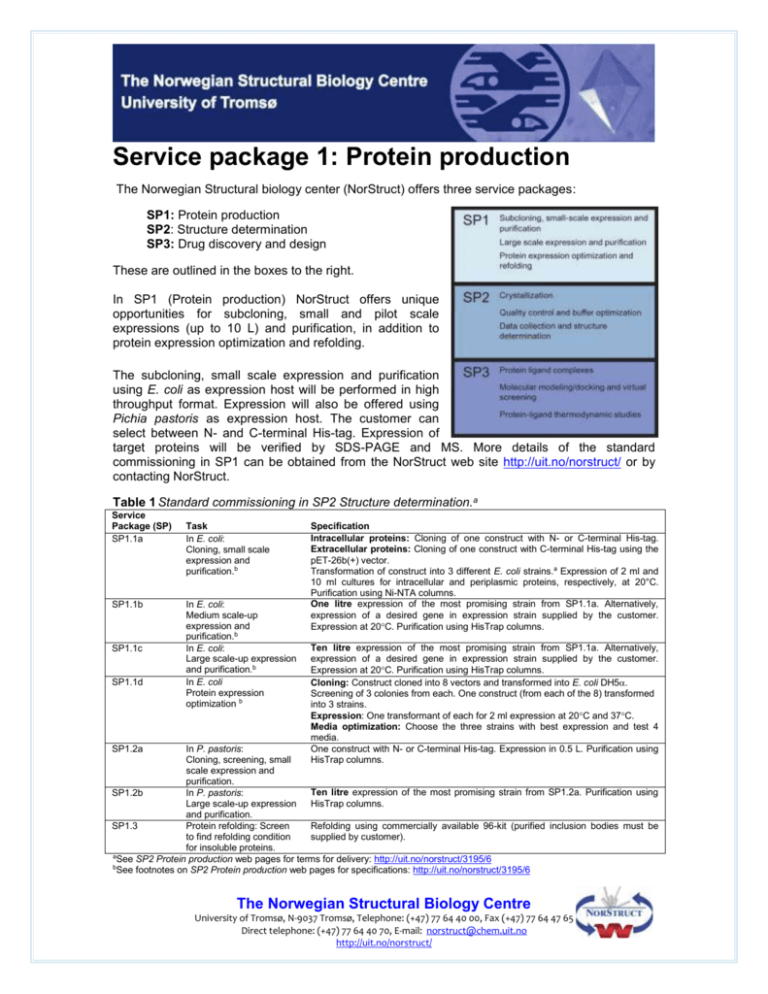
Service package 1: Protein production The Norwegian Structural biology center (NorStruct) offers three service packages: SP1: Protein production SP2: Structure determination SP3: Drug discovery and design These are outlined in the boxes to the right. In SP1 (Protein production) NorStruct offers unique opportunities for subcloning, small and pilot scale expressions (up to 10 L) and purification, in addition to protein expression optimization and refolding. The subcloning, small scale expression and purification using E. coli as expression host will be performed in high throughput format. Expression will also be offered using Pichia pastoris as expression host. The customer can select between N- and C-terminal His-tag. Expression of target proteins will be verified by SDS-PAGE and MS. More details of the standard commissioning in SP1 can be obtained from the NorStruct web site http://uit.no/norstruct/ or by contacting NorStruct. Table 1 Standard commissioning in SP2 Structure determination.a Service Package (SP) SP1.1a SP1.1b SP1.1c SP1.1d SP1.2a Task In E. coli: Cloning, small scale expression and purification.b In E. coli: Medium scale-up expression and purification.b In E. coli: Large scale-up expression and purification.b In E. coli Protein expression optimization b Specification Intracellular proteins: Cloning of one construct with N- or C-terminal His-tag. Extracellular proteins: Cloning of one construct with C-terminal His-tag using the pET-26b(+) vector. Transformation of construct into 3 different E. coli strains.a Expression of 2 ml and 10 ml cultures for intracellular and periplasmic proteins, respectively, at 20°C. Purification using Ni-NTA columns. One litre expression of the most promising strain from SP1.1a. Alternatively, expression of a desired gene in expression strain supplied by the customer. Expression at 20C. Purification using HisTrap columns. Ten litre expression of the most promising strain from SP1.1a. Alternatively, expression of a desired gene in expression strain supplied by the customer. Expression at 20C. Purification using HisTrap columns. Cloning: Construct cloned into 8 vectors and transformed into E. coli DH5. Screening of 3 colonies from each. One construct (from each of the 8) transformed into 3 strains. Expression: One transformant of each for 2 ml expression at 20C and 37C. Media optimization: Choose the three strains with best expression and test 4 media. One construct with N- or C-terminal His-tag. Expression in 0.5 L. Purification using HisTrap columns. In P. pastoris: Cloning, screening, small scale expression and purification. Ten litre expression of the most promising strain from SP1.2a. Purification using SP1.2b In P. pastoris: Large scale-up expression HisTrap columns. and purification. SP1.3 Protein refolding: Screen Refolding using commercially available 96-kit (purified inclusion bodies must be to find refolding condition supplied by customer). for insoluble proteins. aSee SP2 Protein production web pages for terms for delivery: http://uit.no/norstruct/3195/6 bSee footnotes on SP2 Protein production web pages for specifications: http://uit.no/norstruct/3195/6 The Norwegian Structural Biology Centre University of Tromsø, N-9037 Tromsø, Telephone: (+47) 77 64 40 00, Fax (+47) 77 64 47 65 Direct telephone: (+47) 77 64 40 70, E-mail: norstruct@chem.uit.no http://uit.no/norstruct/ Preliminary order form Service package 1: Protein production. Please place tick marks to indicate which tasks are desired. For simplicity this icon can be copied and replaced by the empty boxes. Then return this electronic form by E-mail to NorStruct, and you will receive in response a tailored order form and prizes for placing the final order. NorStruct will not be able to handle genes originating from organisms belonging to risk class 3 or 4. Order: Date: …../…../20…… Title (and NorStruct project number if available): Customer: Institution: Adress: Contact person: E-mail: Phone: Order: (Tick) SP Task SP1.1a In E. coli: Cloning, small scale expression and purification In E. coli: Medium scale-up expression and purification In E. coli: Large scale-up expression and purification. In E. coli Protein expression optimization In P. pastoris: Cloning, screening, small scale expression and purification. In P. pastoris: Large scale-up expression and purification. Protein refolding: Screen to find refolding condition for insoluble proteins. SP1.1b SP1.1c SP1.1d SP1.2a SP1.2b SP1.3 . The Norwegian Structural Biology Centre University of Tromsø, N-9037 Tromsø, Telephone (+47) 77 64 40 00, Fax (+47) 77 64 47 65 Direct telephone (+47) 77 64 40 70, E-mail: norstruct@chem.uit.no http://uit.no/norstruct/ -2- General information Details Information Protein name: Function of protein: protease, DNA binding, etc. ……………….. ………………..………………..……………….. ………………..………………..……………….. DNA details: Number of nucleotides: Sequence: ……………….. Please paste the nucleotide sequence here: Protein details: Number of amino acids: Molecular weight (kDa): GenBank accession number: pI (theoretical or measured): Sequence: ……………….. ……………….. ……………….. ……………….. Please paste the amino acid sequence here: Construct details: Does the construct contain tags: If yes, how many, which type and which order: Does the construct contain TEV cleavage site: Original source organism: Risk class for this sourcea: Risk class of proteina: yes no ………………………………………………………… ………………………………………………………… yes no ………………..……………….. -1; -2; -3; -4 a viral protein; a toxin; a prion protein; a virulence factor; other, no risk. The customer must provide NorStruct with ALL necessary information about the risk classes of the original source and of the produced protein to take all necessary safety precautions, to avoid any harm to the environment and/or people. a NorStruct may not be able to handle genes originating from organisms that belong to risk class 3 or 4. For proteins being viral proteins, toxins, prion proteins or virulence factors, NorStruct might have restrictions handling such proteins. Possible risk factors and the feasibility of handling such high risk class projects will be dealt with in close collaboration with each customer. For more details on the risk classes, please consult the web page (in Norwegian) about protection concerning biological risk factors: http://www.lovdata.no/cgi-wift/ldles?doc=/sf/sf/sf-19971219-1322.html Cloning and small scale expression and purification details Details Information The Norwegian Structural Biology Centre University of Tromsø, N-9037 Tromsø, Telephone (+47) 77 64 40 00, Fax (+47) 77 64 47 65 Direct telephone (+47) 77 64 40 70, E-mail: norstruct@chem.uit.no http://uit.no/norstruct/ -3- Customer provides: Sub-cloned construct Delivery: Vector/system: ……………….. Genomic DNA cDNA (obligatory for eukaryotic proteins) Other details: …………………………………………………………….. Small scale expression type (SP1.1a, SP1.1d, SP1.2a): Intracellular protein in E. coli with: N-Term. His-tag C-Term. His-tag Extracellular protein in E. coli with: C-Term. His-tag Expression in P. pastoris: Protein expression optimization (SP1.1d): Vectors to be used or truncated forms of the construct (up to 8): Strains to be used (up to 3): Temperature to be used: Media to be used: 8 vectors (specify in Additional information section at the end of the form) Truncated forms (please provide details in the Additional information section at the end of the form): ……………….. ……………….. ……………….. ……………….. ……………….. The customer’s suggestion for primer details: Antibodies and or assay: Protein specific antibodies will be provided Activity assay will be provided Other specifications Medium and large scale protein production details Details Information Delivery: Customer provides: Stab culture containing transformed bacterial culture Excised agarose gel containing transformed bacterial culture Other details: …………………………………………………………….. Production type: Medium scale Large scale The Norwegian Structural Biology Centre University of Tromsø, N-9037 Tromsø, Telephone (+47) 77 64 40 00, Fax (+47) 77 64 47 65 Direct telephone (+47) 77 64 40 70, E-mail: norstruct@chem.uit.no http://uit.no/norstruct/ -4- Small scale expression details: Vector and strain: Temperature: Media: Induction details: Proof: Small scale purification details: Chromatographic method (His Trap, ionic exchange, gelfiltration etc.): Temperature: Buffers (type(s) and pH): Proof: Antibodies and or assay: ……………….. ……………….. ……………….. ……………….. ……………….. Please insert picture of SDS gel here (obligatory): Please insert MS results (if available) here: ……………….. ……………….. ……………….. Please insert picture of chromatogram(s) here (obligatory): Please insert picture of SDS gel of purified protein (obligatory): Protein specific antibodies will be provided Activity assay will be provided Other specifications: Additional protein characteristics. In order to obtain soluble protein it is advisable to perform sequence analysis that can aid us in the protein expression and purification stages. We encourage all our customers to perform and report such an analysis. . Factors Question Information Glycosylation 1) 2) Cysteines 1) 2) Metal binding 1) 2) 3) Is glycosylation known for this protein? Is this something to consider? 1) ……………….. Are there free cysteines in this protein (if yes; how many)? Should reducing agents be considered during purification? 1) ……………….. Are there any metal binding sites? If yes, which and how many? (e.g. Fe, Mg, Mn, Zn, etc) Are there any metal binding motifs? Zink fingers, CxxC, [Fe-S], etc? 1) ……………….. 2) ……………….. 2) ……………….. 2) Nol If yes, which reducing agent: -mercaptoethanol DTT Other, specify:…… 3) ……………….. The Norwegian Structural Biology Centre University of Tromsø, N-9037 Tromsø, Telephone (+47) 77 64 40 00, Fax (+47) 77 64 47 65 Direct telephone (+47) 77 64 40 70, E-mail: norstruct@chem.uit.no http://uit.no/norstruct/ -5- Co-factor 1) Could there be any co-factor binding sites; ATP, NAD(P)+, FMN, others? 1) ……………….. If yes, should this be added in to the buffer? Yes No Membrane 1) Is it possible that all or part of the protein could be membrane attached? Could trans-membrane elements be removed to increase the protein solubility? 1) ……………….. 2) 2) ……………….. Signal peptides 1) 2) Are there any signal sequences? Should this possibly be removed? 1) ……………….. 2) ……………….. Domains 1) Are there multiple domains? If yes please specify residue range. 1) ……………….. Oligomerisation 1) Is oligomerisation known for this protein, dimer, trimer etc.? 1) ……………….. Additional information Please provide us with: 1) Construct details for SP1.1d 2) Any additional information about the protein since this can be critical for the success of the cloning, expression and protein purification. Additional details about cloning, expression, purification: Information Additional contact persons In case of NorStruct would like to request more details please provide us with the name of contact person(s) involved in the project that can help us with additional information. Name Address E-mail Phone The Norwegian Structural Biology Centre University of Tromsø, N-9037 Tromsø, Telephone (+47) 77 64 40 00, Fax (+47) 77 64 47 65 Direct telephone (+47) 77 64 40 70, E-mail: norstruct@chem.uit.no http://uit.no/norstruct/ -6-