Molecular microbial ecology
advertisement

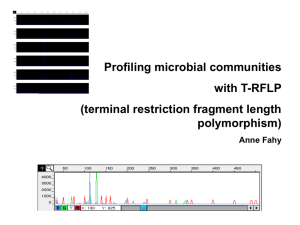
Molecular Microbial Ecology ??? ??? ??? ??? ??? ??? ??? ??? The Challenge for Microbial Ecology Habitat Culturability (%) Seawater 0.001-0.1 0.25 0.25 0.3 Freshwater Sediments Soil How do you study something you can’t grow in the lab? From Amann et al. 1995 Microbiological Reviews The grand picture, from environment to identification Head et al. 1998 A more classical approach Head et al. 1998 Ribosomal RNA (rRNA) •Everybody has it •Contains both highly conserved and variable regions -allows making comparisons between different organisms over long periods of time (evolutionary history) •Not laterally transferred between organisms •Huge and growing database Universal Tree of Life BACTERIA BACTERIA You Are Here EUKARYA EUKARYA ARCHAEA ARCHAEA Primers can be designed to amplify hypervariable regions, but are specific to Eubacteria vs. Archae • 16S rRNA Bacteria primer pairs – Several hypervariable regions • 16S rRNA Archaea primer pairs – Several hypervariable regions Usual procedure in molecular microbial ecology: •Obtain environmental sample (soil, seawater, fresh water, organism such as human gut) •Extract total DNA •PCR amplify and obtain “amplicons” •Or clone DNA, and grow up clones •Genotype/sequence DNA •Characterize microbial diversity Alternative routes for molecular ecological studies in microbiology • Get “community fingerprint” via T-RFLP fingerprint profiles • Get “community fingerprint” via DGGE and sequence bands • Get species identification by – Clone and sequence clones – Skip cloning, go straight into sequencing (massively parallel sequencing, MPS) Alternative routes for molecular ecological studies in microbiology • Get “community fingerprint” via T-RFLP • Get “community fingerprint” via DGGE and sequence bands • Get species identification by – Clone and sequence clones – Skip cloning, go straight into sequencing (massively parallel sequencing, MPS) Denaturing gradient gel electrophosis (DGGE): DNA melts at a certain point What do you do with the sequences? • Perform a similarity search (database) • Align the sequences (common ancestry) • Build a tree (phylogeny and taxonomy) BLAST Basic Local Alignment Search Tool http://blast.ncbi.nlm.nih.gov/Blast.cgi Alignments of sequences Alternative routes for molecular ecological studies in microbiology • Get “community fingerprint” via T-RFLP • Get “community fingerprint” via DGGE and sequence bands • Get species identification by – Clone and sequence clones – Skip cloning, go straight into sequencing (massively parallel sequencing, MPS) • Built clone libraries from deep-sea rocks • Compared them to one another and other habitats 16S RNA sequences Santelli et al. 2008 Community Overlap Santelli et al. 2008 Alternative routes for molecular ecological studies in microbiology • Get “community fingerprint” via T-RFLP • Get “community fingerprint” via DGGE and sequence bands • Get species identification by – Clone and sequence clones – Skip cloning, go straight into sequencing (massively parallel sequencing, MPS) MPS Approaches Schematic courtesy of B. Crump The next generation sequencing methods Platform Million base Cost per Average read pairs per run base (cents) length (base pairs) Dye-terminator (ABI 3730xl) (classic method) 0.07 0.1 700 454-Roche pyrosequencing (next gen.) 400 0.003 400 2,000 0.0007 35 Illumina sequencing (next gen.) From Hugenholtz and Tyson 2008 V3, V6 and V6 hypervariable regions in 16S rRNA genes, and taxon specific conserved primer sites for PCR (%coverage = % species amplified) How many species in 1 L of vent fluid? > 36,000 eubacterial species! ~3,000 archea species Now we know who is there: What next? • Quantify particular groups: FISH or qPCR Head et al. 1998 Fluorescent In-Situ Hybridization (FISH) Schleper et al. 2005 Quantitative (Real Time) PCR • Detection of “amplification-associated fluorescence” at each cycle during PCR • No gel-based analysis • Computer-based analysis • Compare to internal standards • Must insure specific binding of probes/dye Quantitative PCR Primers used to amplify mcrA, an important gene for adaptation to anoxic sediments (note different primers are used to detect different groups) Now we know who and how many: What next? • Metagenomics • RNA-based methods • Many many more… Metagenomics a.k.a., Community Genomics, Environmental Genomics Does not rely on Primers or Probes (apriori knowledge)! Image courtesy of John Heidelberg Metagenomics Access genomes of uncultured microbes: Functional Potential Metabolic Pathways Horizontal Gene Transfer … From the Most “Simple” Microbial Communities… •Acid Mine Drainage (pH ~0!) •Jillian Banfield (UC Berkeley) •Well-studied, defined environment with ~4 dominant members •Were able to reconstruct almost entire community “metagenome” •Tyson et al. 2004 … to the potentially most diverse! Venter et al. 2004 •The Sorcerer II Global Ocean Sampling Expedition •J. Craig Venter Institute “Sequence now, ask questions later” •Very few genomes reconstructed •Sequenced 6.3 billion DNA base pairs (Human genome is ~3.2) from top 5 m of ocean •Discovered more than 6 million genes… and they are only halfway done!