The BCM Microarray Core Facility
advertisement
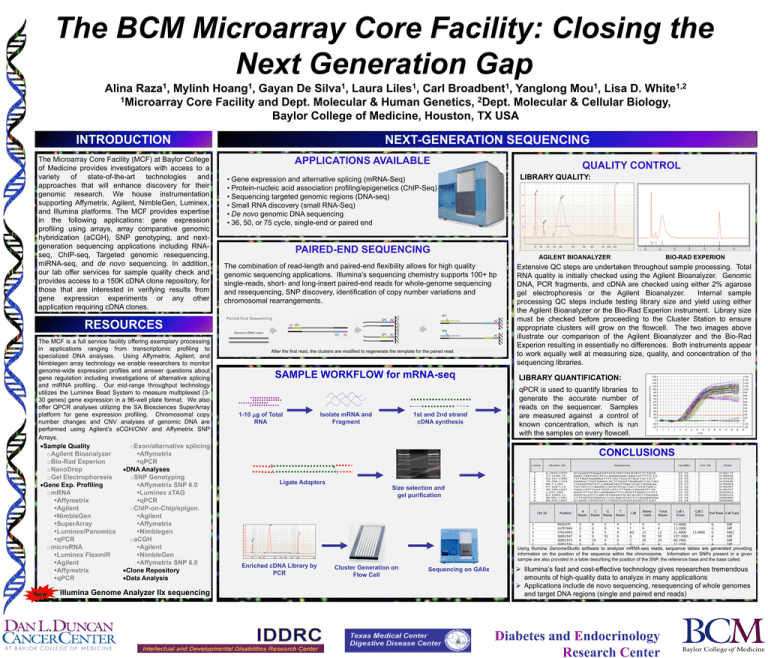
The BCM Microarray Core Facility: Closing the Next Generation Gap 1 Raza , 1 Hoang , 1 Silva , 1 Liles , 1 Broadbent , 1 Mou , 1,2 White Alina Mylinh Gayan De Laura Carl Yanglong Lisa D. 1Microarray Core Facility and Dept. Molecular & Human Genetics, 2Dept. Molecular & Cellular Biology, Baylor College of Medicine, Houston, TX USA INTRODUCTION The Microarray Core Facility (MCF) at Baylor College of Medicine provides investigators with access to a variety of state-of-the-art technologies and approaches that will enhance discovery for their genomic research. We house instrumentation supporting Affymetrix, Agilent, NimbleGen, Luminex, and Illumina platforms. The MCF provides expertise in the following applications: gene expression profiling using arrays, array comparative genomic hybridization (aCGH), SNP genotyping, and nextgeneration sequencing applications including RNAseq, ChIP-seq, Targeted genomic resequencing, miRNA-seq, and de novo sequencing. In addition, our lab offer services for sample quality check and provides access to a 150K cDNA clone repository, for those that are interested in verifying results from gene expression experiments or any other application requiring cDNA clones. NEXT-GENERATION SEQUENCING APPLICATIONS AVAILABLE Sample Quality oAgilent Bioanalyzer oBio-Rad Experion oNanoDrop oGel Electrophoresis Gene Exp. Profiling omRNA Affymetrix Agilent NimbleGen SuperArray Luminex/Panomics qPCR omicroRNA Luminex FlexmiR Agilent Affymetrix qPCR New oExon/alternative splicing Affymetrix qPCR DNA Analyses oSNP Genotyping Affymetrix SNP 6.0 Luminex xTAG qPCR oChIP-on-Chip/epigen. Agilent Affymetrix Nimblegen oaCGH Agilent NimbleGen Affymetrix SNP 6.0 Clone Repository Data Analysis Illumina Genome Analyzer IIx sequencing LIBRARY QUALITY: • Gene expression and alternative splicing (mRNA-Seq) • Protein-nucleic acid association profiling/epigenetics (ChIP-Seq) • Sequencing targeted genomic regions (DNA-seq) • Small RNA discovery (small RNA-Seq) • De novo genomic DNA sequencing • 36, 50, or 75 cycle, single-end or paired end PAIRED-END SEQUENCING AGILENT BIOANALYZER The combination of read-length and paired-end flexibility allows for high quality genomic sequencing applications. Illumina’s sequencing chemistry supports 100+ bp single-reads, short- and long-insert paired-end reads for whole-genome sequencing and resequencing, SNP discovery, identification of copy number variations and chromosomal rearrangements. RESOURCES The MCF is a full service facility offering exemplary processing in applications ranging from transcriptomic profiling to specialized DNA analyses. Using Affymetrix, Agilent, and Nimblegen array technology we enable researchers to monitor genome-wide expression profiles and answer questions about gene regulation including investigations of alternative splicing and miRNA profiling. Our mid-range throughput technology utilizes the Luminex Bead System to measure multiplexed (330 genes) gene expression in a 96-well plate format. We also offer QPCR analyses utilizing the SA Biosciences SuperArray platform for gene expression profiling. Chromosomal copy number changes and CNV analyses of genomic DNA are performed using Agilent’s aCGH/CNV and Affymetrix SNP Arrays. QUALITY CONTROL After the first read, the clusters are modified to regenerate the template for the paired read. SAMPLE WORKFLOW for mRNA-seq 1-10 mg of Total RNA Isolate mRNA and Fragment 1st and 2nd strand cDNA synthesis BIO-RAD EXPERION Extensive QC steps are undertaken throughout sample processing. Total RNA quality is initially checked using the Agilent Bioanalyzer. Genomic DNA, PCR fragments, and cDNA are checked using either 2% agarose gel electrophoresis or the Agilent Bioanalyzer. Internal sample processing QC steps include testing library size and yield using either the Agilent Bioanalyzer or the Bio-Rad Experion instrument. Library size must be checked before proceeding to the Cluster Station to ensure appropriate clusters will grow on the flowcell. The two images above illustrate our comparison of the Agilent Bioanalyzer and the Bio-Rad Experion resulting in essentially no differences. Both instruments appear to work equally well at measuring size, quality, and concentration of the sequencing libraries. LIBRARY QUANTIFICATION: qPCR is used to quantify libraries to generate the accurate number of reads on the sequencer. Samples are measured against a control of known concentration, which is run with the samples on every flowcell. CONCLUSIONS T A A T Ligate Adapters Size selection and gel purification Using Illumina GenomeStudio software to analyzer mRNA-seq reads, sequence tables are generated providing information on the position of the sequence within the chromosome. Information on SNPs present in a given sample are also provided in a table describing the position of the SNP, the reference base and the base called. Enriched cDNA Library by PCR Cluster Generation on Flow Cell Sequencing on GAIIx Illumina’s fast and cost-effective technology gives researches tremendous amounts of high-quality data to analyze in many applications Applications include de novo sequencing, resequencing of whole genomes and target DNA regions (single and paired end reads) Diabetes and Endocrinology Research Center